当前位置:网站首页>CADD course learning (7) -- Simulation of target and small molecule interaction (semi flexible docking autodock)
CADD course learning (7) -- Simulation of target and small molecule interaction (semi flexible docking autodock)
2022-07-05 20:36:00 【Stunned flounder (】
CADD Course study (7)-- Simulate the interaction between the target and small molecules (AutoDock)
Concept :
Molecular docking is through the study of the interaction between ligand small molecules and receptor biological macromolecules , Predict its affinity , An important method to realize structure based drug design .
Theoretical basis :
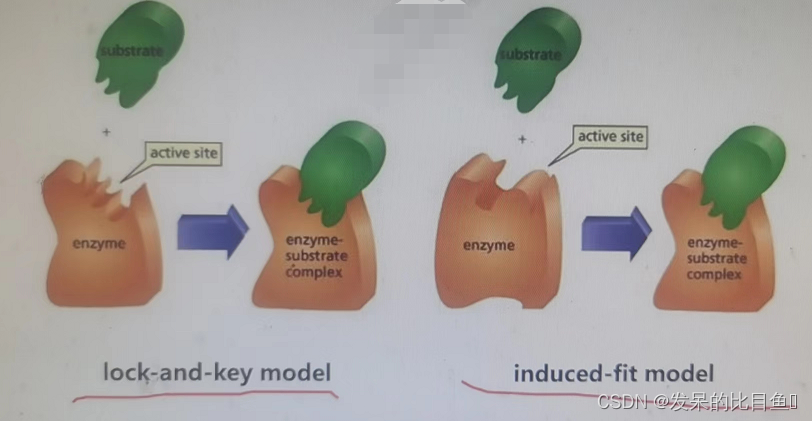
The original idea of molecular docking originated from Fisher E Proposed “ Lock and key model ”, That is, the first condition for the mutual recognition of receptors and ligands is the matching of spatial structure .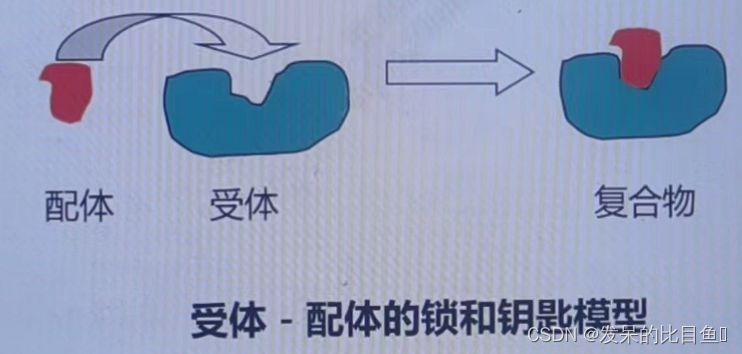
Molecular docking principle
According to geometric matching and energy matching , Find the best interaction mode between two molecules 
Receptor: enzyme, ion channel, antibody……
Ligand: drug, peptide, antigen, polysaccharide…
Receptor-Ligand Interactions:
electrostatic interaction, hydrogen bond, hydrophobic interaction
Docking software
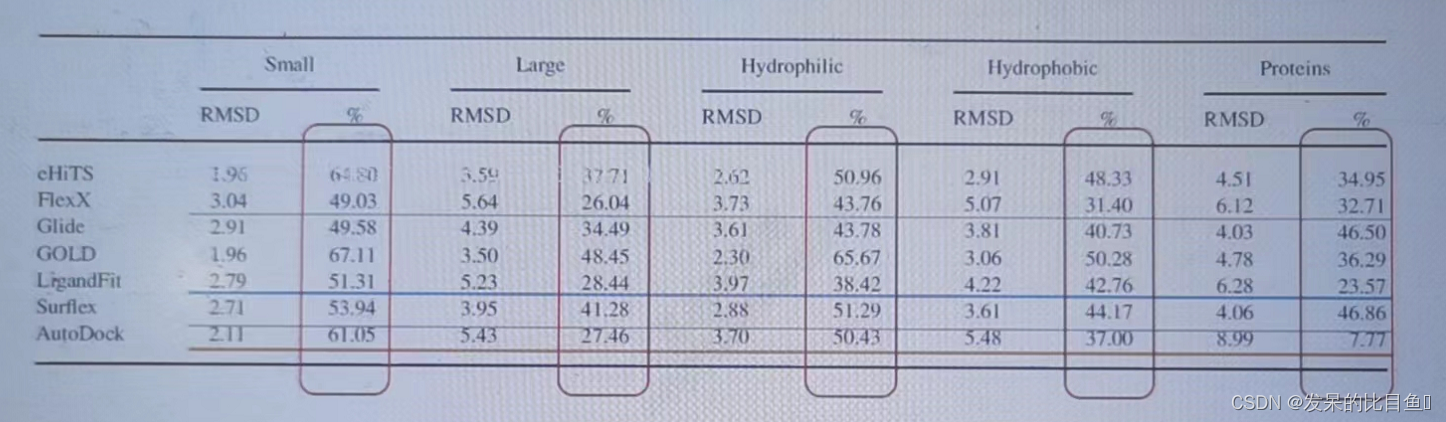
- Different systems have different results ;
- DS(LigandFit),SYBYL(flexx,surflex),AutoDock The results are not much different
- Autodock Good performance , And open source for free , Safe to use
Test set :1300 protein-ligands complexes from PDBbind database
The general process of molecular docking
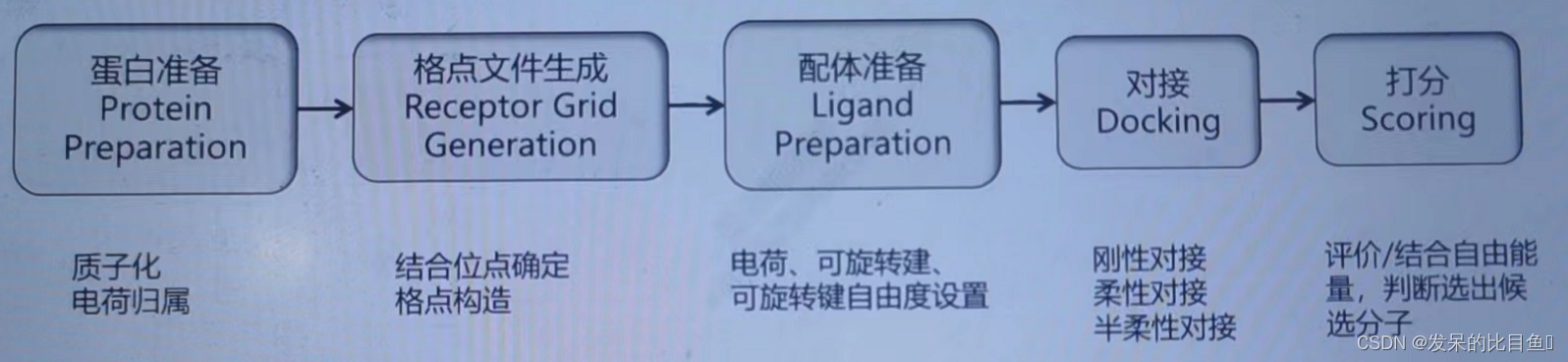
principle : When the ligand binds to the receptor , There is electrostatic interaction with each other 、 Hydrogen bond interaction 、 Van der Waals force interaction and hydrophobic force . The binding of ligand and receptor must meet the principle of mutual matching , That is, the geometry of ligand and receptor is complementary 、 Electrostatic interaction complementary matching 、 Hydrogen bond interaction complementary matching 、 Hydrophobic interactions complement each other .
classification
- Rigid butt joint : During docking , The conformation of the study system does not change . Suitable for investigating relatively large systems , Such as the docking between proteins and between proteins and nucleic acids .
- Semi flexible docking : During docking , The conformation of the studied system, especially the ligand, is allowed to change within a certain range . It is suitable for docking between macromolecules and small molecules .
- Flexible docking : During docking , The conformation of the research system is basically free to change . It is generally used to accurately consider the recognition between molecules , Because the conformation of the system can change during the calculation , So the calculation cost the most .
AutoDock: Small molecule compounds - Protein docking software
http://autodock.scripps.edu
install
- install Python Running environment , And set environment variables
- Autodock***,exe(windows edition ) Double clicking will decompress three files :autodock4.exe、autogrid4.exe and cygwin1.dl, And automatically copy them to C:\Windows\System32\ Under the folder , installation is complete . then ,DOS You can directly enter “autodock4” or
“autogrid4” function . For example, manually copy the above three files to other directories , Each time you call this command, you need to point out the path of the storage directory - Mgltool**,exe(windows edition ): Just like installing an ordinary program, double-click to install .
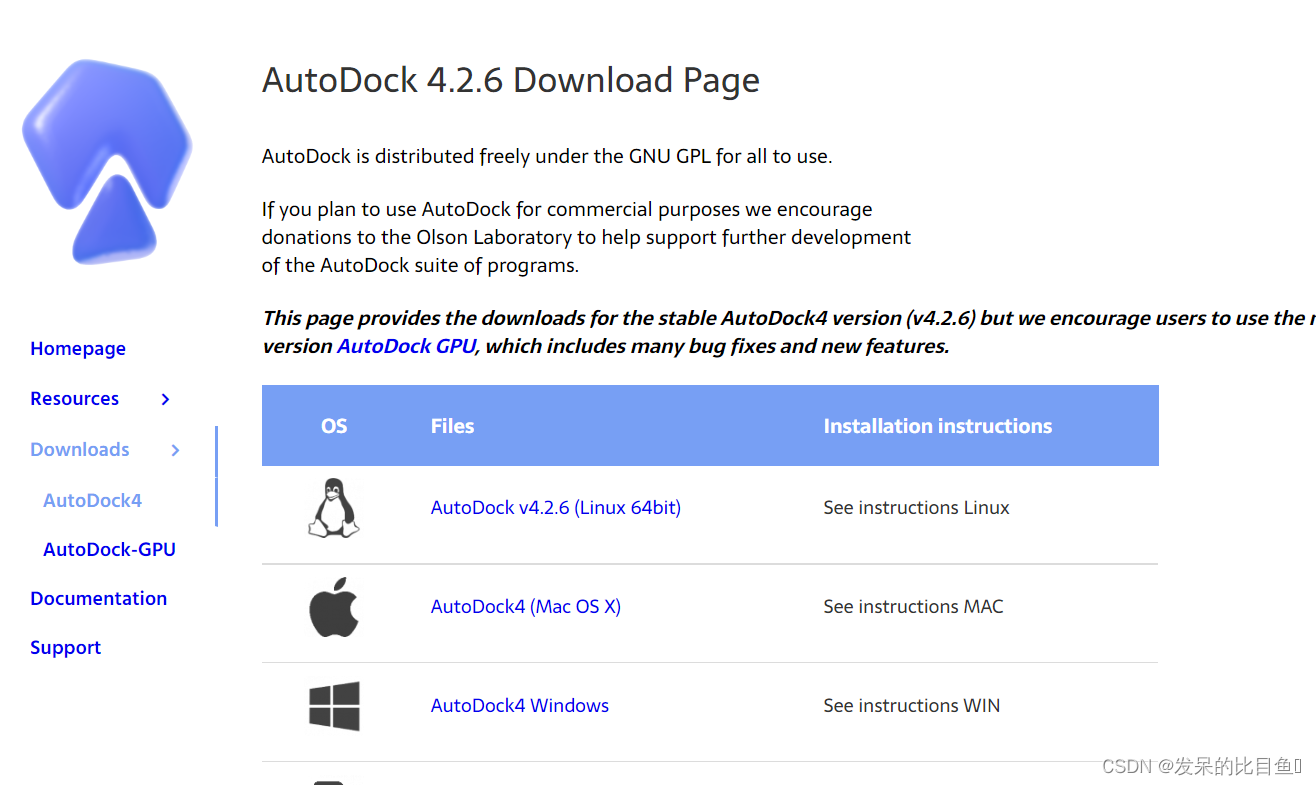


Docking cases
AutoDock Docking system :PDB 1IEP
AutoDock Docking steps :
- Prepare ligand and receptor files (AutoDockTools)
- Prepare grid file (AutoGrid)
- Molecular docking calculations (AutoDock)

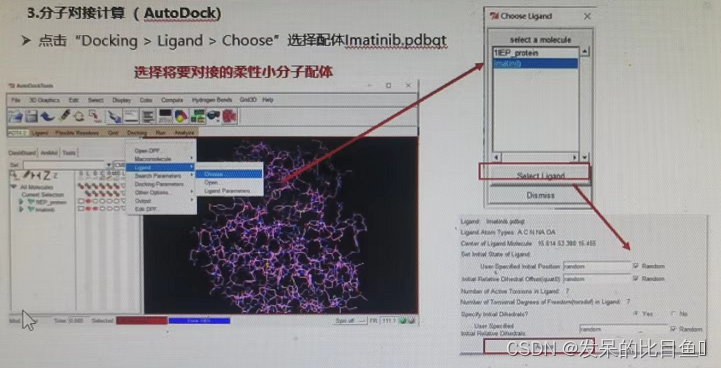
1. Prepare ligand receptor file (AutoDockTools)
(1)Pymo download 11EP, Delete B chain , Remove water molecules ,CL ion
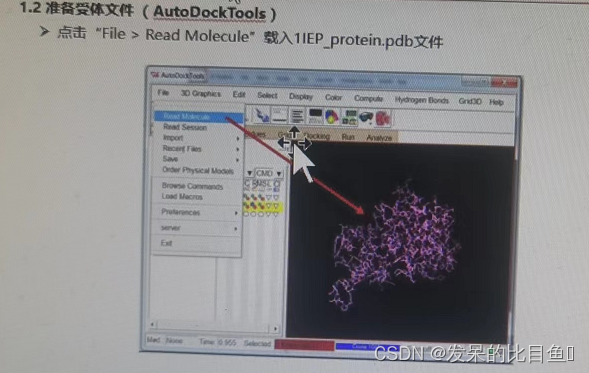
(2) Select the protein part ( Does not contain ligands STI), Save as 1EP_protein.pdb
(3) Select ligand (STI), Save as lmatinib.pdb notes :PDB 1IEP The ligands of ST That is to say lmatinib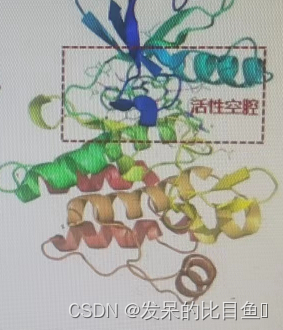
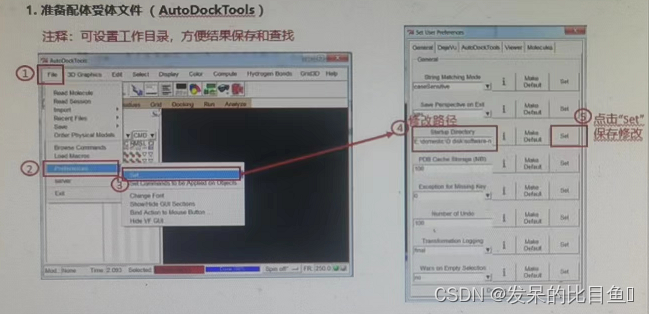
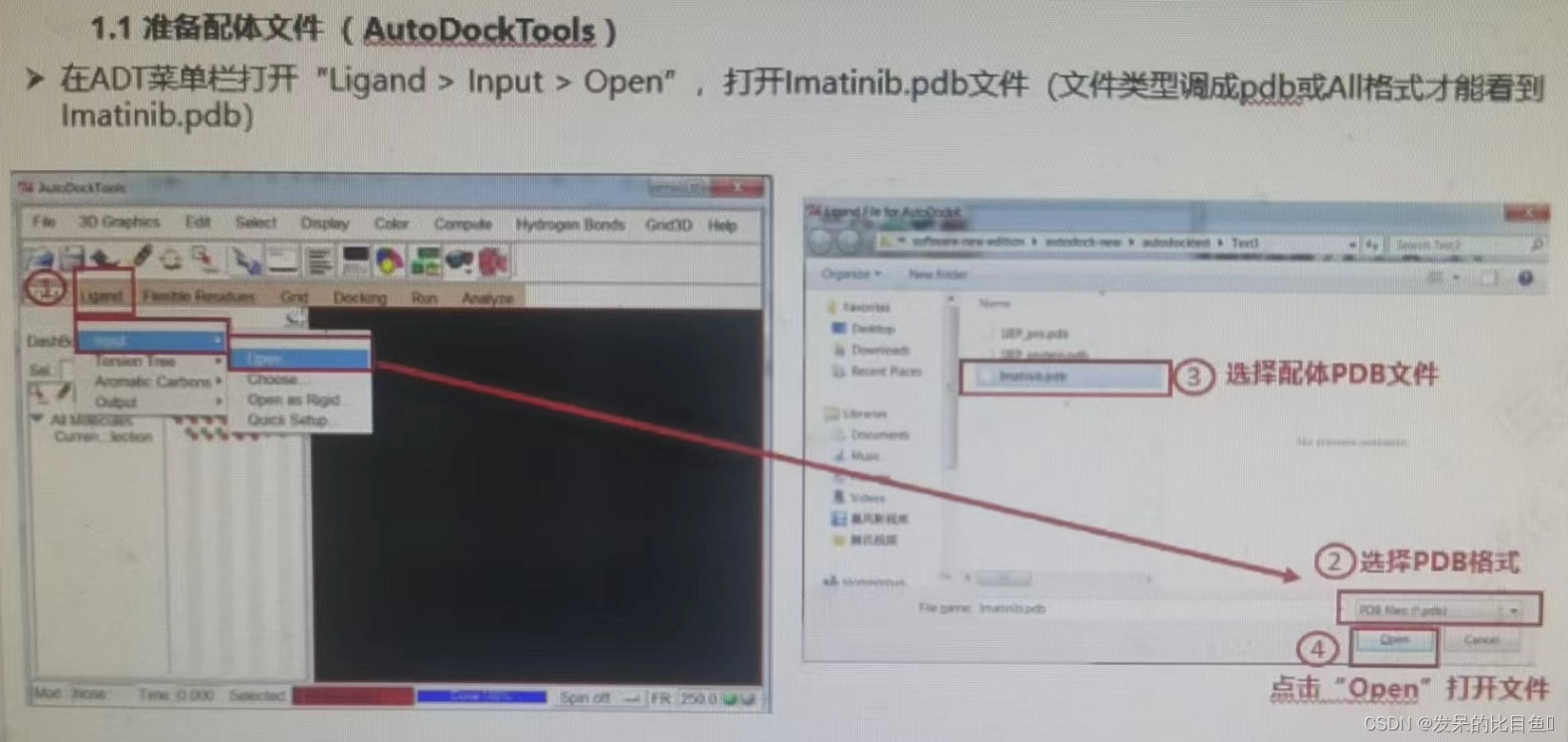
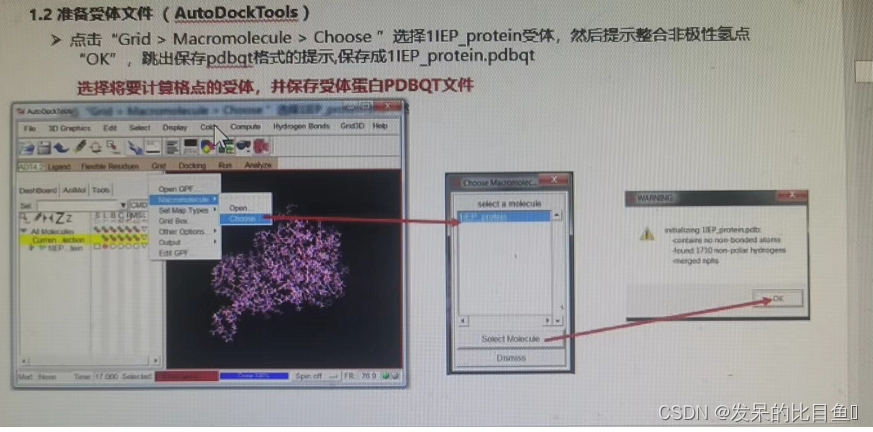

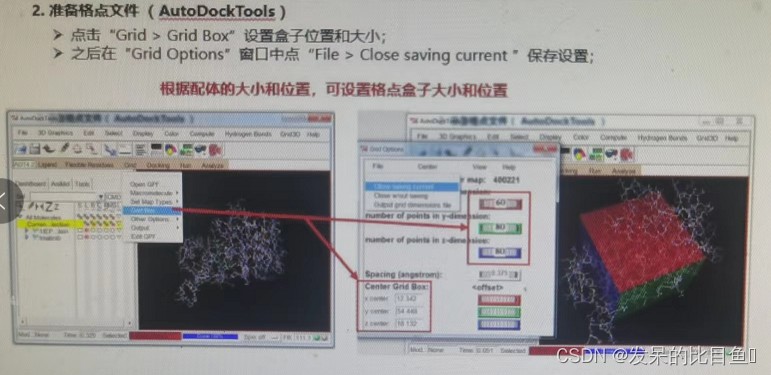
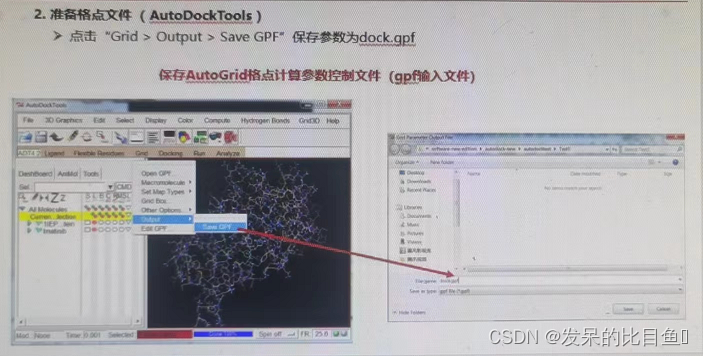
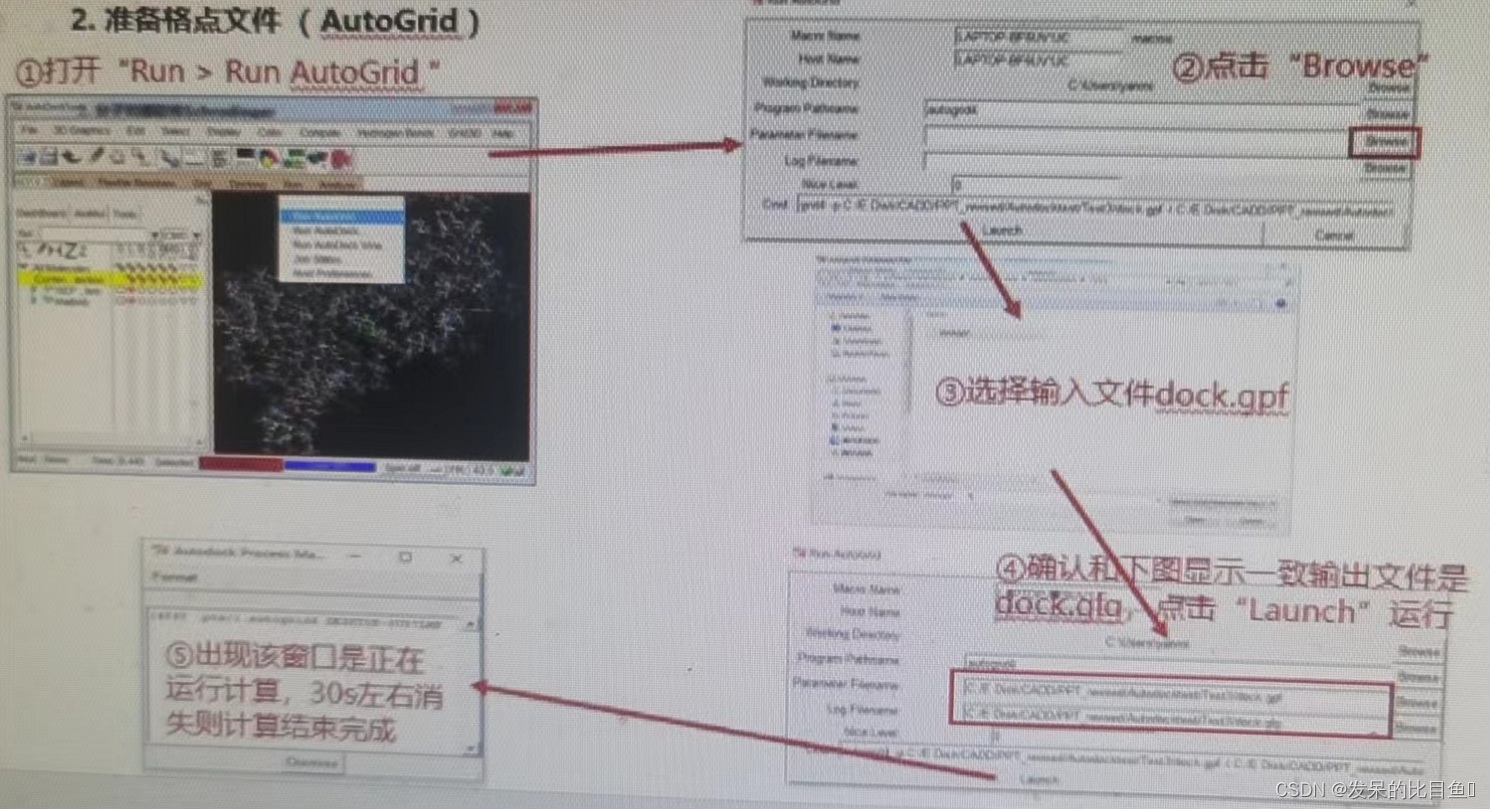
If the interface cannot RUN, You can use the command line :.\autogrid4.exe -p .\dock.gpf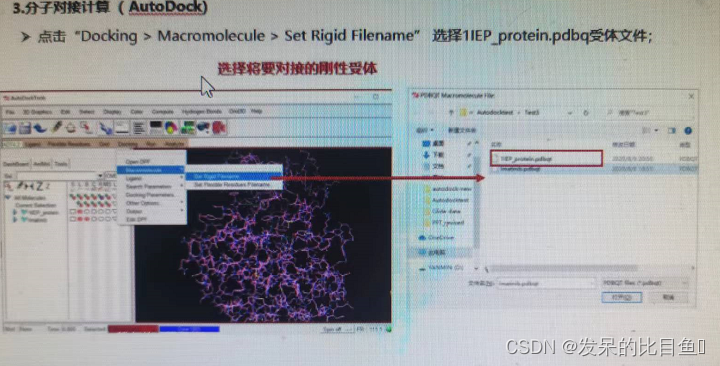
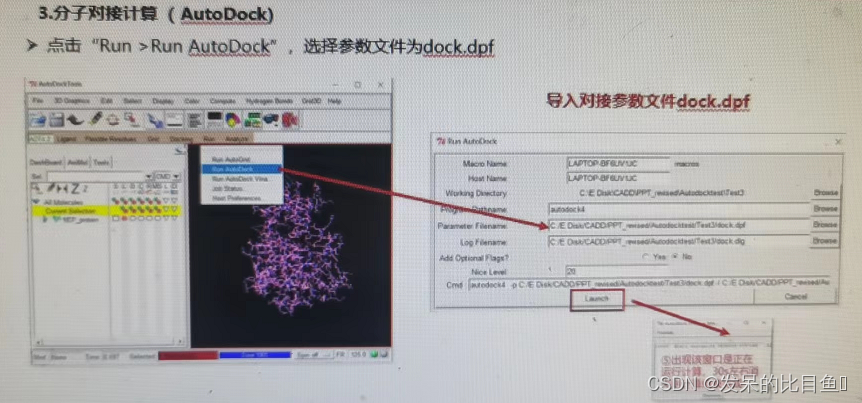
If not RUN, Using the command line .\autodock4.exe -p .\dock.dpf
Summary of semi flexible docking
AutoDock Docking system :PDB 1lEP AutoDock Half docking steps :
- Prepare ligand and receptor files (AutoDockTools)
Autodock Tools Ligand file preparation :
① Add H atom 、 Add gasteiger Empirical charge 、 Combine nonpolar hydrogen
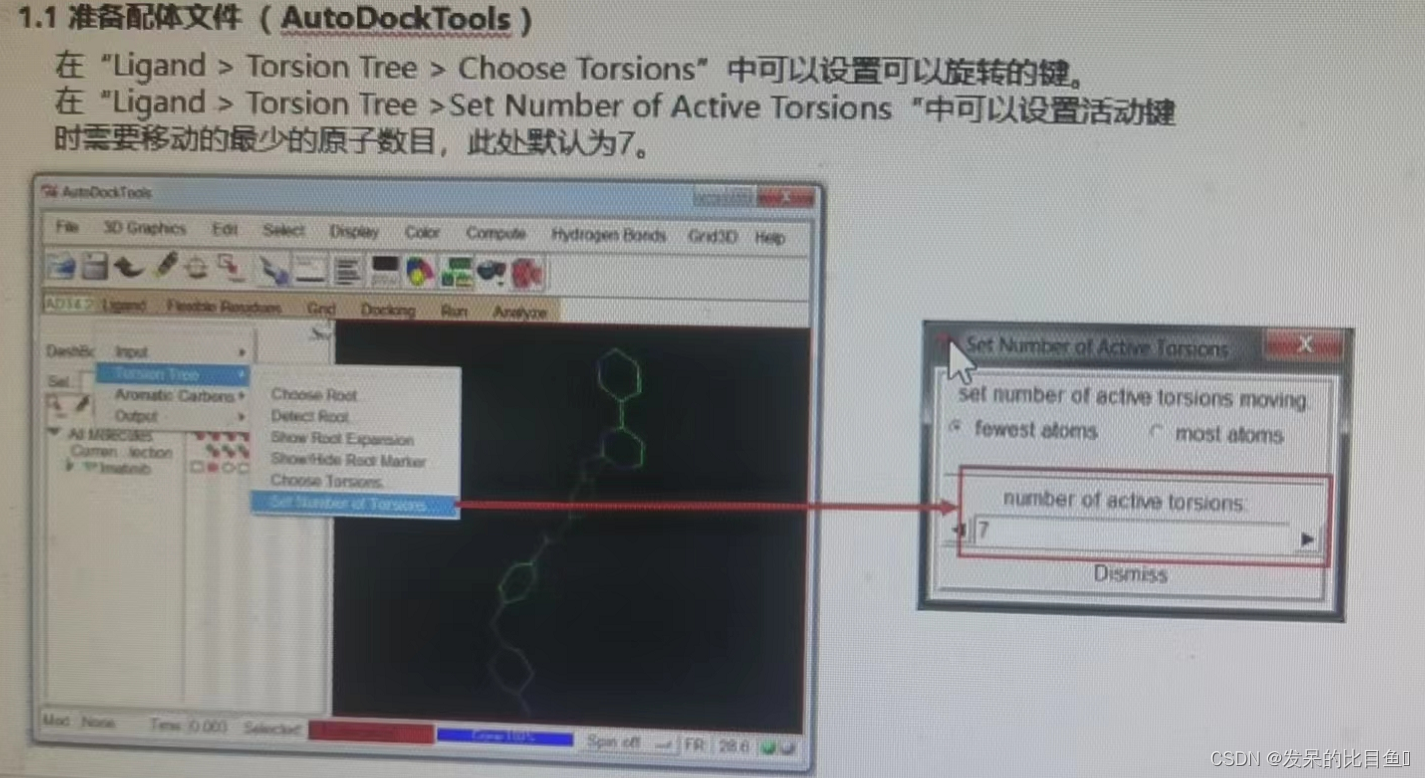
② Set the rotatable key ;
③ preservation lig.pdbqt;
Autodock Tools Receptor document preparation :
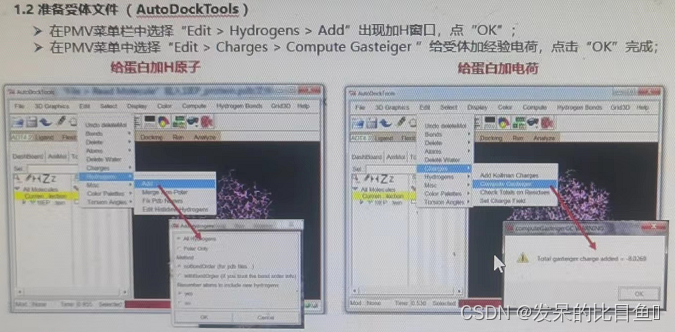
① Delete water 、 Add H atom 、 Add gasteiger Empirical charge 、 Merge non
② preservation rec.pdbqt; - Prepare grid file (AutoGrid)
① Selective rigid receptor ;
② Set the probe atom type 、 Set the size and position of the box ;
③ preservation dock.gpf Parameter control file
④ function AutoGrid Program calculation grid ; - Molecular docking calculations (AutoDock)
① Choose rigid receptors and flexible ligands ;
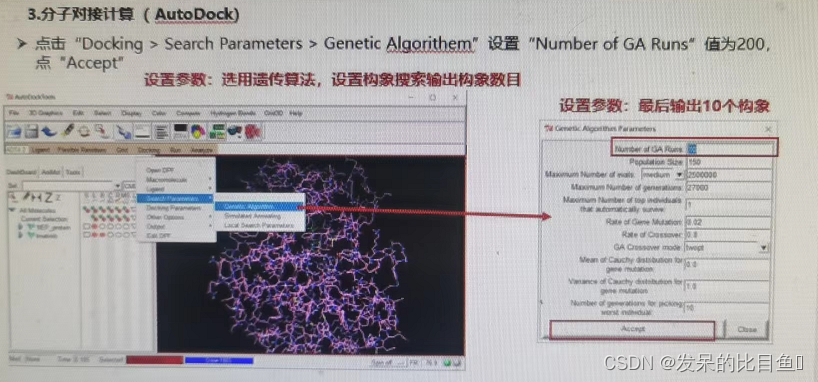
② Set the number of output conformations of genetic algorithm conformation search ;
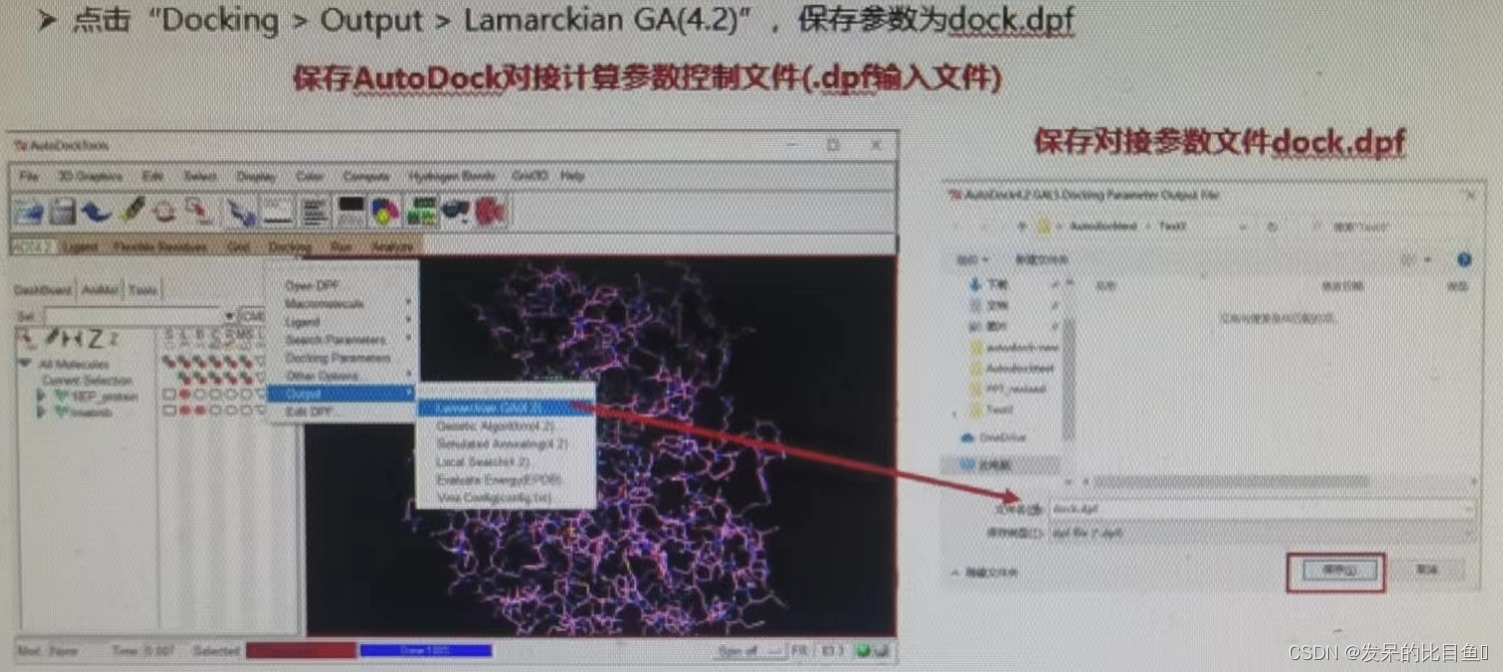
③ preservation dock.dpf Parameter control file
④ function AutoDock Program docking , Output dock.dlg;
AutoDock Docking system : PDB 1IEP
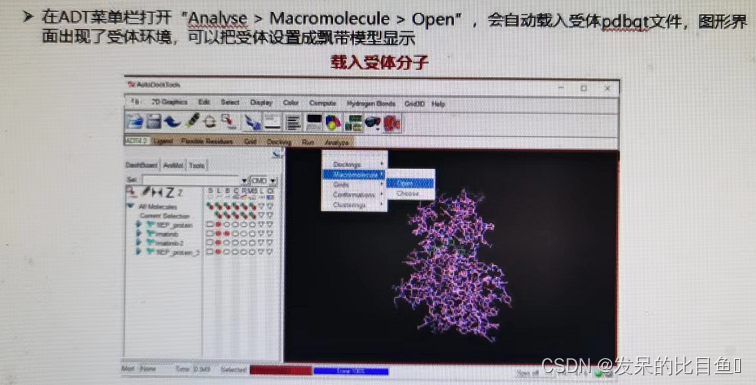
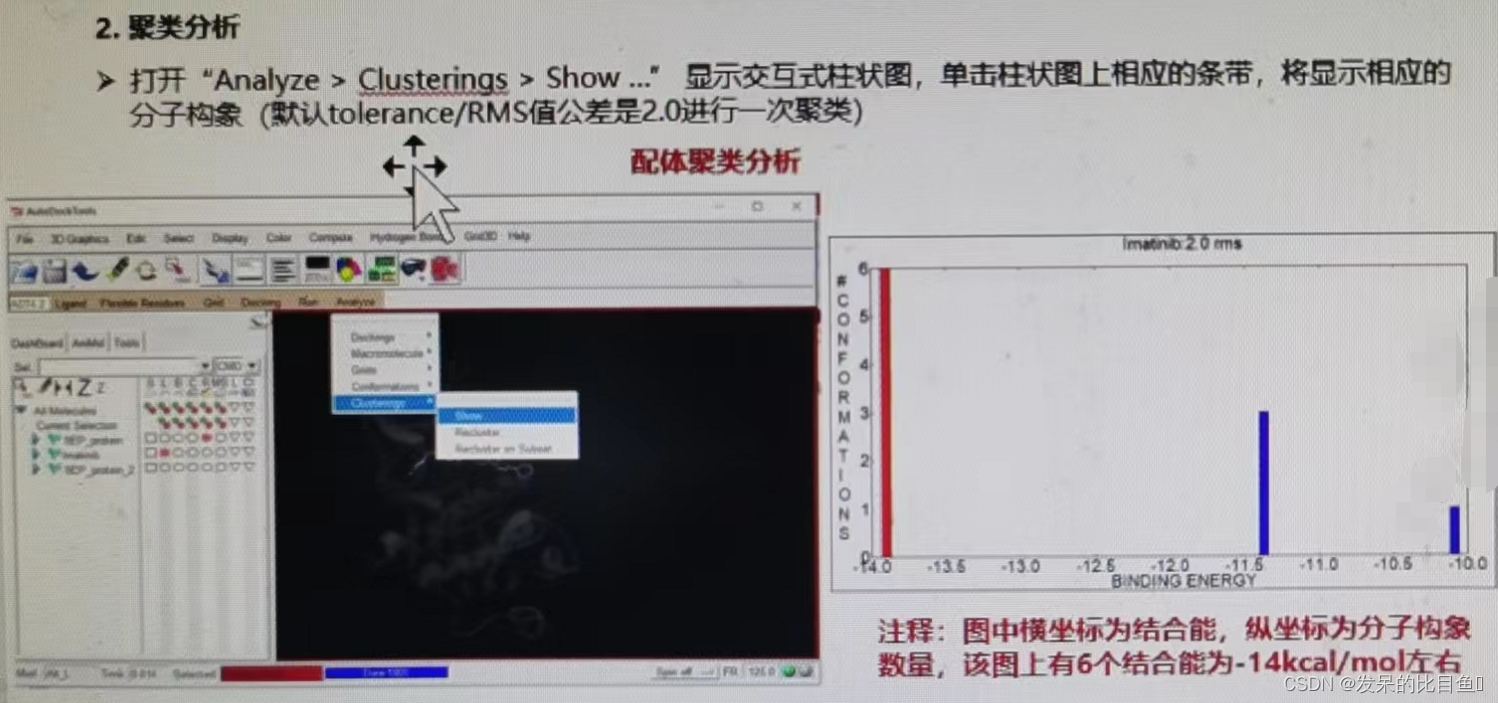
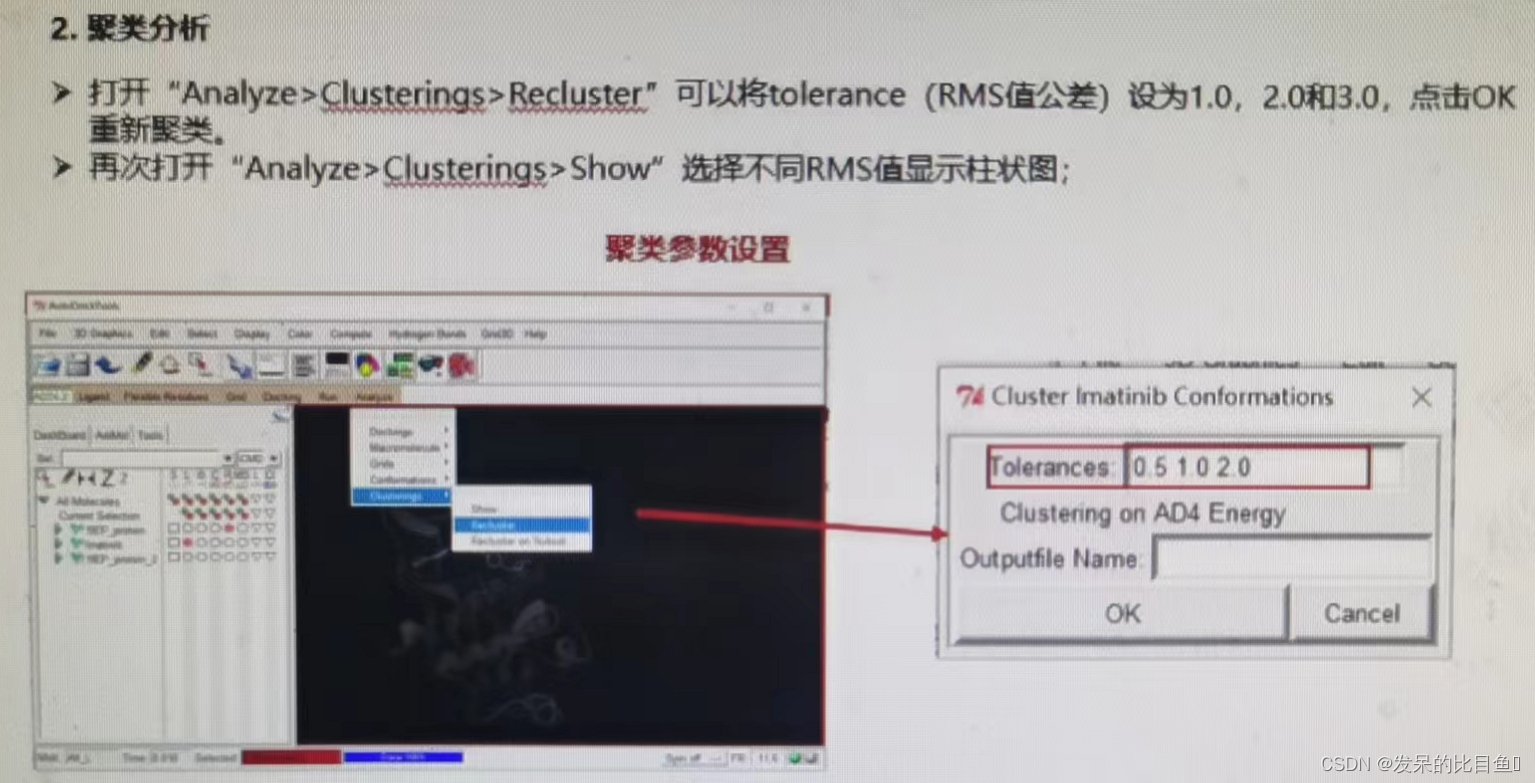
AutoDock Analysis and evaluation of docking results :
- Energy analysis
- Clustering analysis
- And X-ray Crystal conformation comparison
- Choice of optimal binding conformation

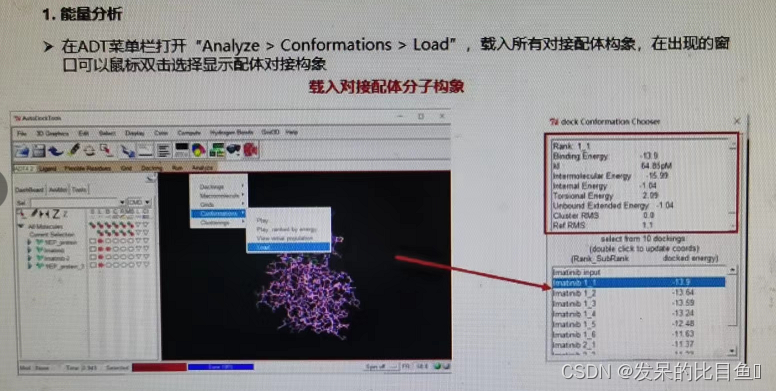
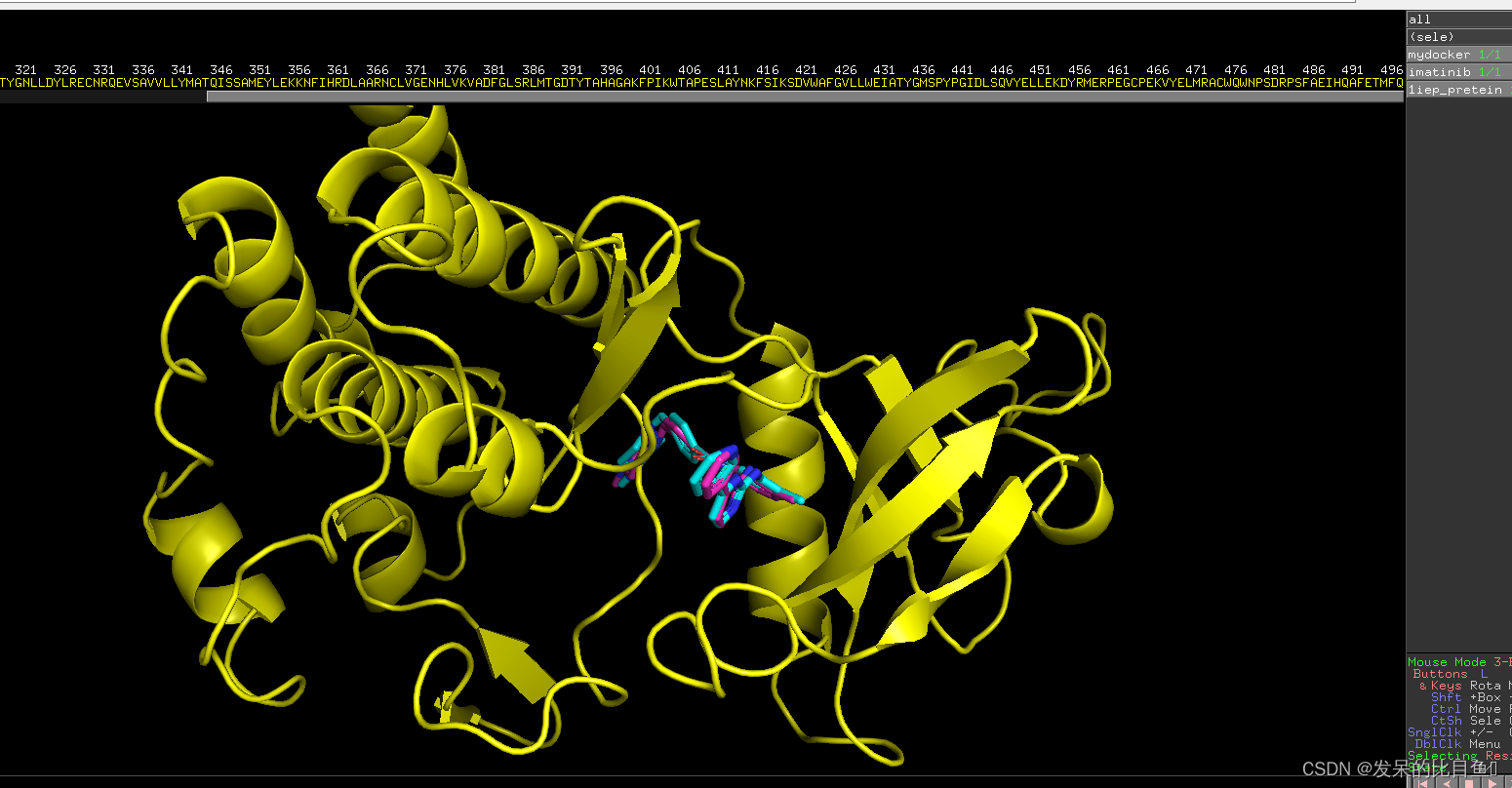
The molecular conformations and complexes of docking ligands are derived
- Click on “Buld current” Two docking conformations can be superimposed for comparison ;
- Click on “Build Al" All ligand conformations can be superimposed ( If the number is large, don't try );
- Click on “Play Parameters” You can set the playback frame rate 、 Start frame 、 End frame, etc
- Click on “Write current” Save the currently displayed docking ligand conformation ; Click on Write Complex Save the currently displayed docking receptor - Ligand complex conformation .

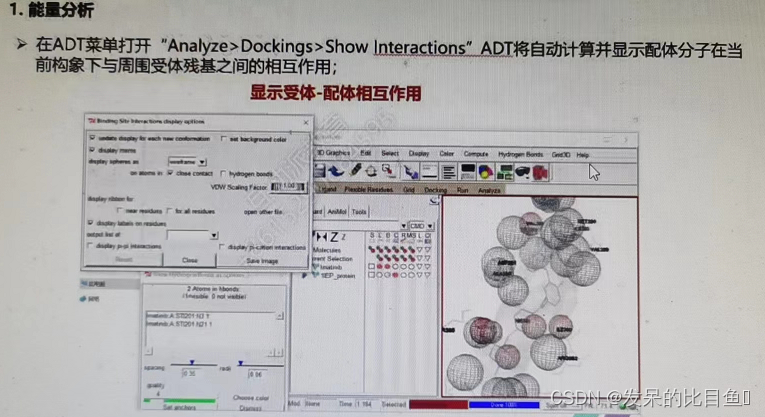
stay PyMol see
summary ( Selection principle of optimal binding conformation )
Choice of optimal binding conformation
- Take the measured crystal structure as a reference ;( If there are receptor ligand binding crystals )
- Choose the one with high affinity in the active pocket ;
- Choose the one where clustering is dominant ;
- Recipient - Ligand noncovalent interaction analysis ( Combined with the literature, judge and screen the appropriate conformation )
- Fully flexible docking to obtain a better conformation ;
- The result of semi flexible docking ( compound ) Conduct molecular dynamics simulation to achieve steady state , Docking again in a steady-state conformation ;
边栏推荐
- Informatics Olympiad 1337: [example 3-2] word search tree | Luogu p5755 [noi2000] word search tree
- [UE4] unrealinsight obtains the real machine performance test report
- Practical demonstration: how can the production research team efficiently build the requirements workflow?
- CVPR 2022 | 常见3D损坏和数据增强
- Duchefa丨MS培养基含维生素说明书
- 资源道具化
- 小程序页面导航
- Applet global configuration
- Abbkine BCA法 蛋白质定量试剂盒说明书
- Scala basics [HelloWorld code parsing, variables and identifiers]
猜你喜欢
![[quick start of Digital IC Verification] 2. Through an example of SOC project, understand the architecture of SOC and explore the design process of digital system](/img/1d/22bf47bfa30b9bdc2e8fd348180f49.png)
[quick start of Digital IC Verification] 2. Through an example of SOC project, understand the architecture of SOC and explore the design process of digital system

CVPR 2022 | common 3D damage and data enhancement
ProSci LAG-3 重组蛋白说明书
Leetcode brush question: binary tree 13 (the same tree)

kubernetes资源对象介绍及常用命令(五)-(ConfigMap&Secret)

Abnova丨荧光染料 620-M 链霉亲和素方案
IC popular science article: those things about Eco

2022北京眼睛健康用品展,护眼产品展,中国眼博会11月举办
Fundamentals - configuration file analysis

Duchefa细胞分裂素丨二氢玉米素 (DHZ)说明书
随机推荐
Sort and projection
Rainbow 5.7.1 supports docking with multiple public clouds and clusters for abnormal alarms
科普|英语不好对NPDP考试有影响吗 ?
CCPC 2021威海 - G. Shinyruo and KFC(组合数,小技巧)
欢迎来战,赢取丰厚奖金:Code Golf 代码高尔夫挑战赛正式启动
Simple understanding of interpolation search
Leetcode skimming: binary tree 16 (path sum)
信息学奥赛一本通 1340:【例3-5】扩展二叉树
如何形成规范的接口文档
Informatics Olympiad 1337: [example 3-2] word search tree | Luogu p5755 [noi2000] word search tree
信息学奥赛一本通 1337:【例3-2】单词查找树 | 洛谷 P5755 [NOI2000] 单词查找树
【数字IC验证快速入门】8、数字IC中的典型电路及其对应的Verilog描述方法
[quick start to digital IC Verification] 8. Typical circuits in digital ICs and their corresponding Verilog description methods
Abnova丨 MaxPab 小鼠源多克隆抗体解决方案
常用的视图容器类组件
Applet event binding
Ros2 topic [01]: installing ros2 on win10
Duchefa丨P1001植物琼脂中英文说明书
3.3、项目评估
When JS method passes long type ID value, precision loss will occur