当前位置:网站首页>Interpretation of the paper: "bert4bitter: a basic model for improving bitter peptide prediction based on transformer (BERT) bidirectional encoder representation"
Interpretation of the paper: "bert4bitter: a basic model for improving bitter peptide prediction based on transformer (BERT) bidirectional encoder representation"
2022-07-23 12:21:00 【Windy Street】
BERT4Bitter: a bidirectional encoder representations from transformers(BERT)- based model for improving the prediction of bitter peptides
Article address :https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/btab133/6151716
DOI:https://doi.org/10.1093/bioinformatics/btab133
Periodical :Bioinformatics(2 District )
Influencing factors :6.937
Release time :2021 year 2 month 26 Japan
data :https://github.com/Shoombuatong/Dataset-Code/tree/master/iBitter
The server :http://pmlab.pythonanywhere.com/BERT4Bitter
1. An overview of the article
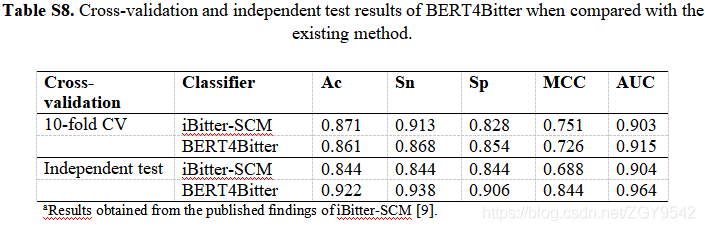
There are a large number of new peptide sequences available in the post genome era , The development of automatic computational models for identifying novel bitter peptides is very necessary . Proposed by the author BERT4Bitter Is based on the converter (BERT) The two-way encoder of represents , Used to predict bitter peptides directly from amino acid sequences , Without using any structural information , This is the first time to adopt based on BERT Model to identify bitter peptides . Compared with widely used machine learning models ,BERT4Bitter Of ACC The cross validation achieved 0.861, In the independent test, we achieved 0.922. On separate datasets BERT4Bitter Better than existing methods ,ACC and MCC Improved respectively 8.0% and 16.0%.
2. Preface
Many drugs have bitter taste and strong effort , Designed to improve the bitter taste , So as to improve the compliance of drug intake , The development of rapid and accurate identification tools for predicting bitter peptides represents a fundamental component in drug development and nutrition research . iBitter-SCM It is based on the recording card method (SCM) Predictor of , Peptides directly from amino acid sequences can be predicted , Use 20 Amino acids and 400 Dipeptide propensity score . iBitter-SCM In cross validation and independent testing ACC Respectively reached 0.871 and 0.844.
Sequence based predictors , Use different feature coding methods to build , For example, based on the characteristics of composition , Combine the characteristics of the transformation distribution and the characteristics based on physical and chemical properties . First , Compared with a single feature descriptor , The combination of various feature descriptors can effectively improve the prediction performance . But using a combination of various features will include redundant and noisy information , This leads to poor prediction results . secondly , The problem of high-dimensional feature space can be overcome by identifying informative features through feature selection algorithm , But this process is very regular , Because it requires multiple manual , Tedious, trial and attack . In the method based on deep learning , Based on natural language processing (NLP) Technology's raw data automatically generates features , Without systematic design and selection of feature coding .
In this study , Each peptide sequence is based on NLP Technology is seen as a sentence , among 20 Each of the amino acids is considered a word . Specially ,BERT4Bitter Pass the peptide sequence as input to BERT Method , Used to automatically generate feature descriptors , Without systematic design and selection of feature coding . Besides , The author also introduces the use of NLP Heuristic feature coding method , namely TF-IDF,Pep2Vec and FastText, Used to represent peptide sequences , Then as a feature as input to machine learning (ML) Model of [ Decision tree (DT) , Extreme random tree (Etree),K a near neighbor (knn), Support vector machine (SVC), Logical regression (LR), Multilayer perceptron (MLP), Naive Bayes (NB), Random forests (RF), Support vector machine (SVM) And extreme gradient boost (XGB)]. For predicting bitter peptides BERT4Bitter The schematic framework of is shown in Figure 1 Shown in .BERT4Bitter Its advantages and main contributions can be summarized as follows :
- Use the original peptide sequence , There is no need to systematically design and select feature codes .
- And amino acid composition (AAC), Amino acid index (AAI), Dipeptide composition (DPC), Pseudo amino acid composition (PseAAC) And tripeptide composition (TPC) Compared with .
- be based on BERT Model of BERT4Bitter Better than existing methods ( Machine learning models evaluated by independent test sets ).
- Developed Web The server .
3. data
Benchmark data set BTP640 By non redundant 320 Bitter peptide and 320 A non bitter peptide , Press 8:2 Proportion is randomly divided into training sets (BTP-CV ) And independent test sets (BTP-TS).BTP-CV The data set contains 256 Bitter peptides and 256 A non bitter peptide , and BTP-TS The data set contains 64 Bitter peptides and 64 A non bitter peptide .
4. Method
4.1 be based on NLP Feature coding for
be based on NLP The method has been successfully applied to several fields across drug discovery ,NLP The method is to automatically treat the original input data as a set of resolvable functions ( Provide the function of original input data ). This paper adopts Pep2Vec and FastText Formal NLP The concept of transforming peptide sequences into N- Word carrier of dimension , Where the word means 20 A naturally occurring amino acid .TFIDF The method is used to calculate each amino acid from the peptide sequence to N- In the carrier of dimension .
4.1.1 TFIDF
Word frequency - Reverse file frequency (TFIDF) Is based on NLP Information retrieval technology in , Is one of the most commonly used document representation methods .TFIDF By including word frequency (TF) And reverse file frequency (IDF) It consists of two main parts , among TF Represents a given document J(DJ) The words in I(TI) Number of occurrences of , and IDF It refers to the interest of reverse file frequency .
TF-IDF(term frequency–inverse document frequency, Word frequency - Reverse file frequency ) It's a kind of information retrieval (information retrieval) And text mining (text mining) Commonly used weighting techniques .
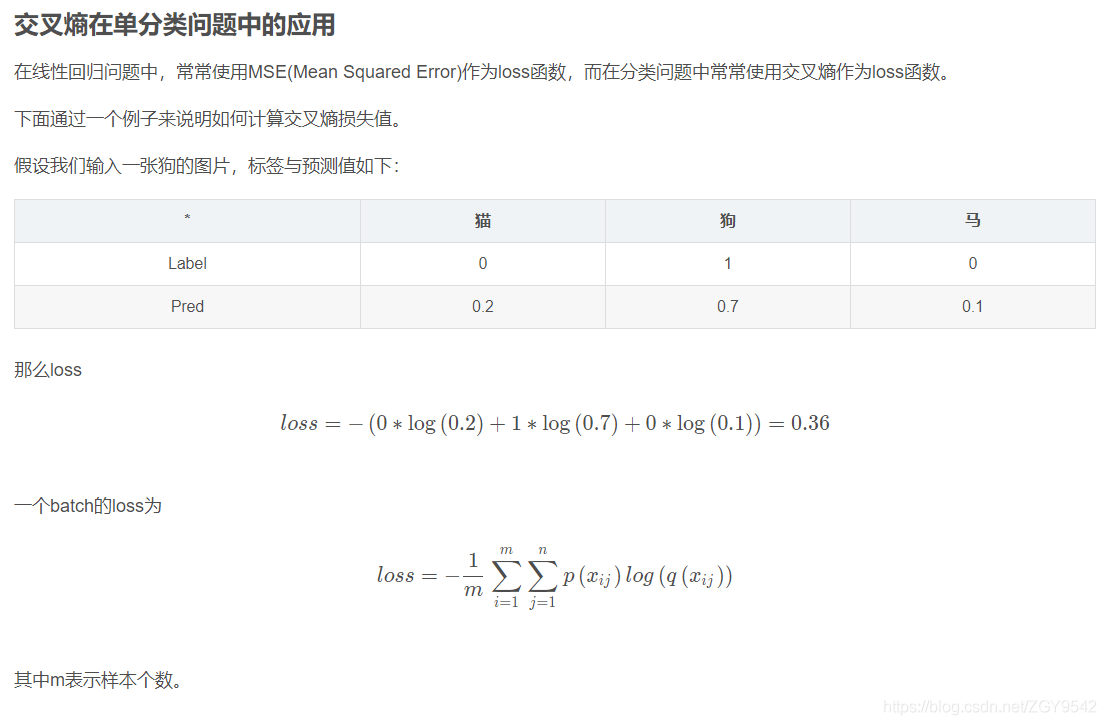
TF-IDF It's a statistical method , To evaluate the importance of a word to a document set or one of the documents in a corpus . The importance of a word increases in proportion to the number of times it appears in the document , But at the same time, it will decrease inversely with the frequency of its occurrence in the corpus .
TF-IDF The main idea of : If the frequency of a word in an article TF high , And it's rarely seen in other articles , It is believed that this word or phrase has a good ability of classification , Suitable for classification .
4.1.2 Pep2Vec
Pep2Vec It's based on Word2Vec Technology for generating biological sequence features ,2016 year ,Aggarwala and Voight A window method is introduced , You can divide the sequence by K-gram To represent each sequence as N Dimension word embedding vector , This can be captured using Hierarchical learning Biophysical and biochemical properties obtained by the process . stay Word2Vec There are two main neural architectures for generating word embedding vectors in : Continuous bag model (CBOW) and Skip-gram.
CBOW Is the context of the known current word , To predict the current word , and Skip-gram On the contrary , When the current word is known , Predict its context .
4.1.3 FastText
Usually , Each peptide sequence is regarded as a sentence containing many words , Each word consists of a bag of characters n-gram form .FastText Method to use words by inserting these special symbols in the boundary , This information can improve the discrimination between prefixes and suffixes from other character sequences .
and CBOW equally ,fastText There are only three layers of models : Input layer 、 Hidden layer 、 Output layer (Hierarchical Softmax), The inputs are all vector words , The output is a specific target, The hidden layer is the average of multiple word vectors . The difference is ,CBOW The input of is the context of the target word ,fastText The input of is multiple words and n-gram features , These features are used to represent a single document ;CBOW The input word of is onehot Encoded ,fastText The input feature of is embedding too ;CBOW The output of is the target vocabulary ,fastText The output of is the class mark corresponding to the document .
It is worth noting that ,fastText On input , Put the character level of the word n-gram Vector as an extra feature ; At output time ,fastText It's layered Softmax, It greatly reduces the training time of the model .
4.2 Deep learning
The author adopts three popular and powerful NLP inspire DL Method , namely BERT, Convolutional neural networks (CNN) And long and short term memory networks (LSTM), Use test sets (BTP-CV) Data set development prediction model .
4.2.1 CNN
be based on CNN The model of includes a total of six layers : An embedded layer , One 1D Convolution layer , A global maximum pool layer , Two Dense Layer and a Dropout layer . The input size of the embedded layer is 21(20 Amino acids and filling space ), One dimensional convolution has 128 Filter 、 The convolution kernel size is 5、 The activation function is Relu. In order to reduce the dimension of the original data set and compress the number of samples , Before applying two fully connected layers, the default 1D Global maximum pool layer . The first fully connected layer contains 50 With Relu Active hidden nodes , Added Dropout by 0.02 Of Dropout Layer prevents the model from over fitting . The second fully connected layer contains 1 Hidden nodes , With binary classification of models Sigmoid Activate . For high performance , At the same time, it provides low generalization error ,Adam Algorithm Used as optimizer , The cross entropy loss method is used as the loss function .
4.2.2 LSTM
be based on LSTM The model of contains five layers : An embedded layer , One LSTM layer , Two fully connected layers and one Dropout layer . be based on LSTM The architecture of the model is similar to that based on CNN Model of , There are only two exceptions :LSTM-based Mold use has 50 Unit LSTM Layer built , Eliminates an unnecessary 1D Global maximum pool layer .
4.2.3 BERT
be based on BERT Our model is a NLP Inspired deep learning methods , It has and is based on CNN The model can effectively capture more global context information than the global receiving domain . In this study , By using traditional BERT The architecture is based on BERT Model of , Its architecture is similar to LSTM Model of , There are only two exceptions : This paper develops BERT-based Model contains 12 layer , The weight of its pre training is used as the embedding layer ; Use with 50 Two way of units LSTM Layer substitution LSTM layer .
4.2.4 Classifier of conventional machine learning
Machine learning methods :DT,Etree,KNN,LR,MLP,NB,RF,SVC,SVM and XGB.
Decision tree (DT) , Extreme random tree (Etree),K a near neighbor (knn), Support vector machine (SVC), Logical regression (LR), Multilayer perceptron (MLP), Naive Bayes (NB), Random forests (RF), Support vector machine (SVM) And extreme gradient boost (XGB)
features :AAC,AAI,DPC,PseAAC and TPC
By using 10 The grid search process of multiple cross validation scheme is used to adopt ML The best prediction accuracy and super parameters of the algorithm ( In the supplementary table S1 The search scope is displayed in ). Last , Use the parameter set with the highest performance for development ML classifier .DT,KNN and NB The classifier is implemented by using its default parameters .
5. Results and discussions
5.1 Analyze the properties of amino acids and dipeptides
Adopted GINI coefficient (MDGI) Average reduced value to rank and estimate each AAC and DPC The importance of features , Supplementary table S2 Lists bitter and non bitter peptides 20 Percentage of amino acids and their use BTP-CV Two classes obtained in the prediction model of the data set and their MDGI Amino acid composition difference between values . Besides , chart 2 Provides a display DPC Heat map of feature importance .

5.2 Different NLP Performance of feature coding
Use ten well-known ML Algorithm ( namely KNN,DT,ETREE,LR,MLP,NB,RF,SVC,SVM, Evaluate three in pairs NLP Heuristic feature coding ( namely FastText,Pep2Vec and TFIDF), adopt 10 Times cross validation and independent testing to evaluate their prediction results .


stay FastText,Pep2Vec and TFIDF In the method , Divide all peptide sequences into groups for use 1-gram,2-gram and 3-gram 's words . Performance comparison shows that ,TFIDF,PEP2VEC and FastText Methods use 1-gram Produce better prediction performance of bitter peptides ( Supplementary table S5). therefore , Each peptide sequence consists of 100 Dimensional vectors are represented and then used for training ML classifier .
5.3 Different NLP Based on the performance of deep learning method

5.4 Bert4Bitter Comparison with other machine learning algorithms



5.5 Bert4bitter Compared with the existing methods

Supplementary knowledge
1. Article to read BERT( Principles ):https://blog.csdn.net/sunhua93/article/details/102764783
2.Keras BERT:https://github.com/CyberZHG/keras-bert/blob/master/README.zh-CN.md
3.tensorflow 2.0 Version based on BERT Tokenizer Text classification :https://zhuanlan.zhihu.com/p/188684911
边栏推荐
- NLP自然语言处理-机器学习和自然语言处理介绍(一)
- 论文解读:《Deep-4mcw2v: 基于序列的预测器用于识别大肠桿菌中的 N4- 甲基胞嘧啶(4mC)位点》
- 单片机学习笔记6--中断系统(基于百问网STM32F103系列教程)
- 把LVGL所有控件整合到一个工程中展示(LVGL6.0版本)
- CPC client installation tutorial
- Deep learning neural network
- Interpretation of the paper: using attention mechanism to improve the identification of N6 methyladenine sites in DNA
- ARM架构与编程3--按键控制LED(基于百问网ARM架构与编程教程视频)
- 编码器的一点理解
- Gartner research: how is China's digital development compared with the world level? Can high-performance computing dominate?
猜你喜欢

编码器的一点理解

Use pyod to detect outliers

时间序列的数据分析(一):主要成分

论文解读:《开发和验证深度学习系统对黄斑裂孔的病因进行分类并预测解剖结果》

2021信息科学Top10发展态势。深度学习?卷积神经网络?

NLP natural language processing - Introduction to machine learning and natural language processing (I)

Gartner research: how is China's digital development compared with the world level? Can high-performance computing dominate?

利用google or-tools 求解数独难题

数据分析(二)

论文解读:《提高N7-甲基鸟苷(m7G)位点预测性能的迭代特征表示方法》
随机推荐
ARM架构与编程5--gcc与Makefile(基于百问网ARM架构与编程教程视频)
2021信息科学Top10发展态势。深度学习?卷积神经网络?
Using or tools to solve the path planning problem with capacity constraints (CVRP)
绿色数据中心:风冷GPU服务器和水冷GPU服务器综合分析
ARM架构与编程1--LED闪烁(基于百问网ARM架构与编程教程视频)
Importance of data analysis
Using pycaret: low code, automated machine learning framework to solve classification problems
Interpretation of yolov3 key code
CPC客户端的安装教程
Smart pointer shared_ PTR and unique_ ptr
CPC client installation tutorial
Numpy summary
g2o安装路径记录--为了卸载
Using Google or tools to solve logical problems: Zebra problem
实用卷积相关trick
Data analysis (I)
谈谈转动惯量
How to develop the liquid cooled GPU server in the data center under the "east to West calculation"?
Notes | (station B) Adult Liu: pytorch deep learning practice (code detailed notes, suitable for zero Foundation)
Matplotlib Usage Summary