当前位置:网站首页>MIT-6874-Deep Learning in the Life Sciences Week5
MIT-6874-Deep Learning in the Life Sciences Week5
2022-06-30 10:09:00 【Wooden girl】
Lecture 6: Regulatory genomicsGene regulation, chromatin accessibility, DNA regulatory code
- AI Application in regulatory omics
- 1 Fundamentals of Biology : Regulatory building blocks
- 2 Traditional methods of regulatory omics and motif discovery
- Insert picture description here
- 3 Basic application of convolutional neural network in transcriptional regulation
- 4 CNNS/RNNs Practical application in regulatory omics : Various architectures
- 4.1 DeepBind: learn motifs, use in(shallow) fully-connected layer, mutations impact
- 4.2 DeepSea: Train model directly on mutational impact prediction
- 4.3 Baseet: Multi-task DNase prediction in 164 cell types, reuse/learn motifs
- 4.4 ChromPuter: Multi-task prediction of different TFs, reuse partner motifs
- 5 Lecture : Stanford Anshul Kundaje,Deep Learning for Reg. Genomics
AI Application in regulatory omics
1 Fundamentals of Biology : Regulatory building blocks
1.1 Gene regulation :Cell diversity, Epigenomics, Regulators(TFs), Motifs, Disease role
Genes are regulated , Express different cell types , So as to produce different organs in the human body .
DNA packaging The meaning of :DNA Highly spiralized , Realize the purpose of chromatin assembly
dna After forming a double chain by double helix , It binds to histones to form nucleosomes , In the formation of supercoiles by continuous spiraling and curling , Finally, they do not short aggregate to form chromosomes / Chromatin . If this kind of highly dense structure is not available, the space required will be very large , It also hinders various metabolic reactions in cells 
(DNA What are the main steps of compression ? - Ryan Answer - You know
https://www.zhihu.com/question/381570484/answer/1103039550)
Nucleosome assembly is the first step in chromosome assembly ,DNA Packed into nucleosomes , About compressed 7 times .
Chromatin is packed and compressed gradually with nucleosome as its basic structure , the 30nm Chromatin fibers 、 Super spiral ring 、 Finally, it is compressed and packaged into chromosomes , A total of four levels of packaging .
● DNA To the nucleosome
● From nucleosome to solenoid (solenoid)
● From solenoid to super solenoid (supersolenoid)
● From super solenoids to chromosomes
DNA Compression in cells is very important .DNA Inner covenant 147 Base pairs loop Form nucleosome (nucleosome), Each nucleosome consists of four Histone form , Probably 50 Nucleosomes form a chromatin filament (chromatin fiber)

Epigenomics – Programming methods for each cell type in the body , By adjusting the compression specific DNA Characteristics of sequence promoter region .
Starter :RNA Polymerase can bind to , Concurrent transcription DNA Part of . The area where cell expression begins to activate
Transcription factor utilization DNA Binding domains identify specific genes in the genome DNA Sequence
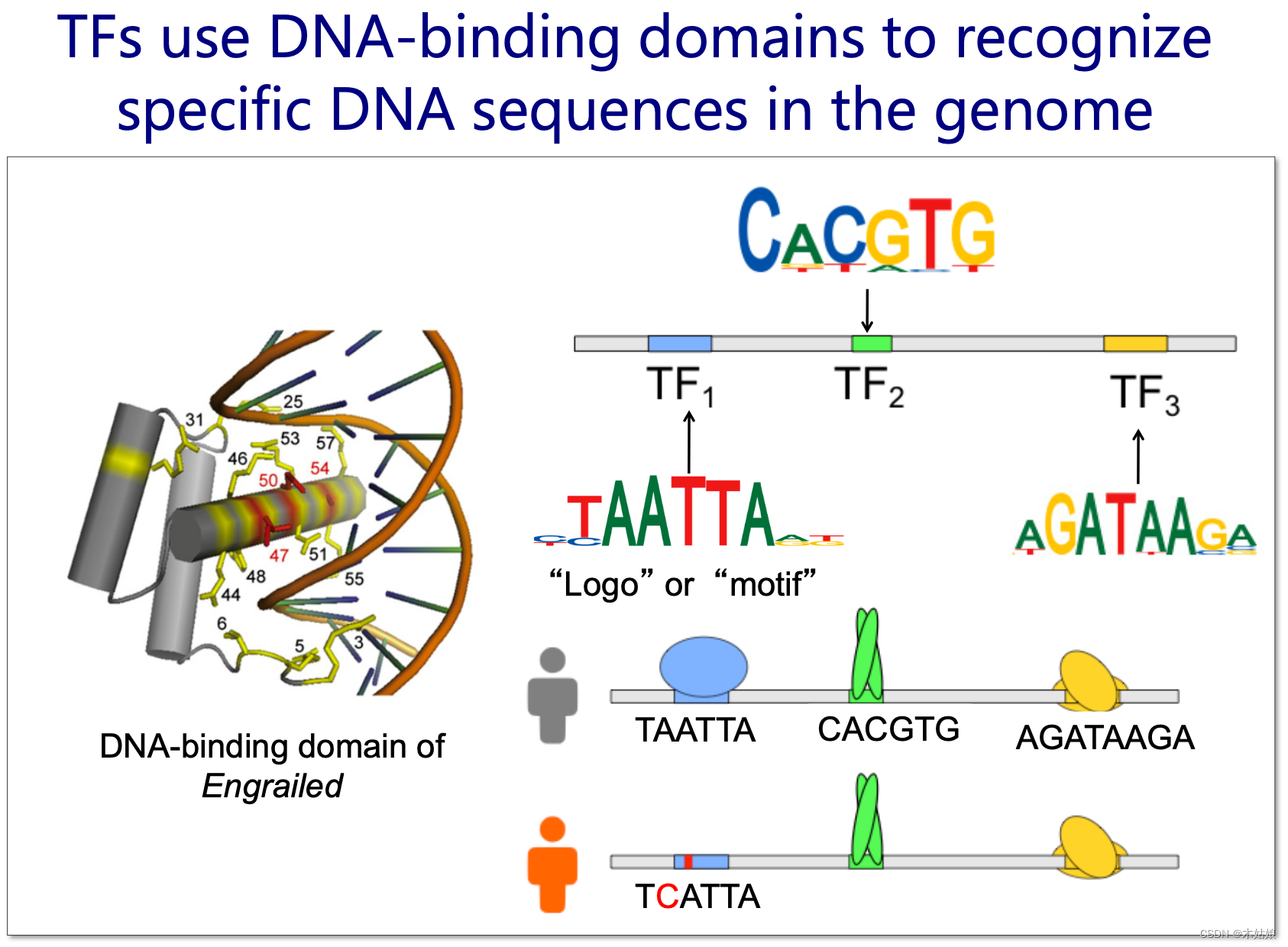
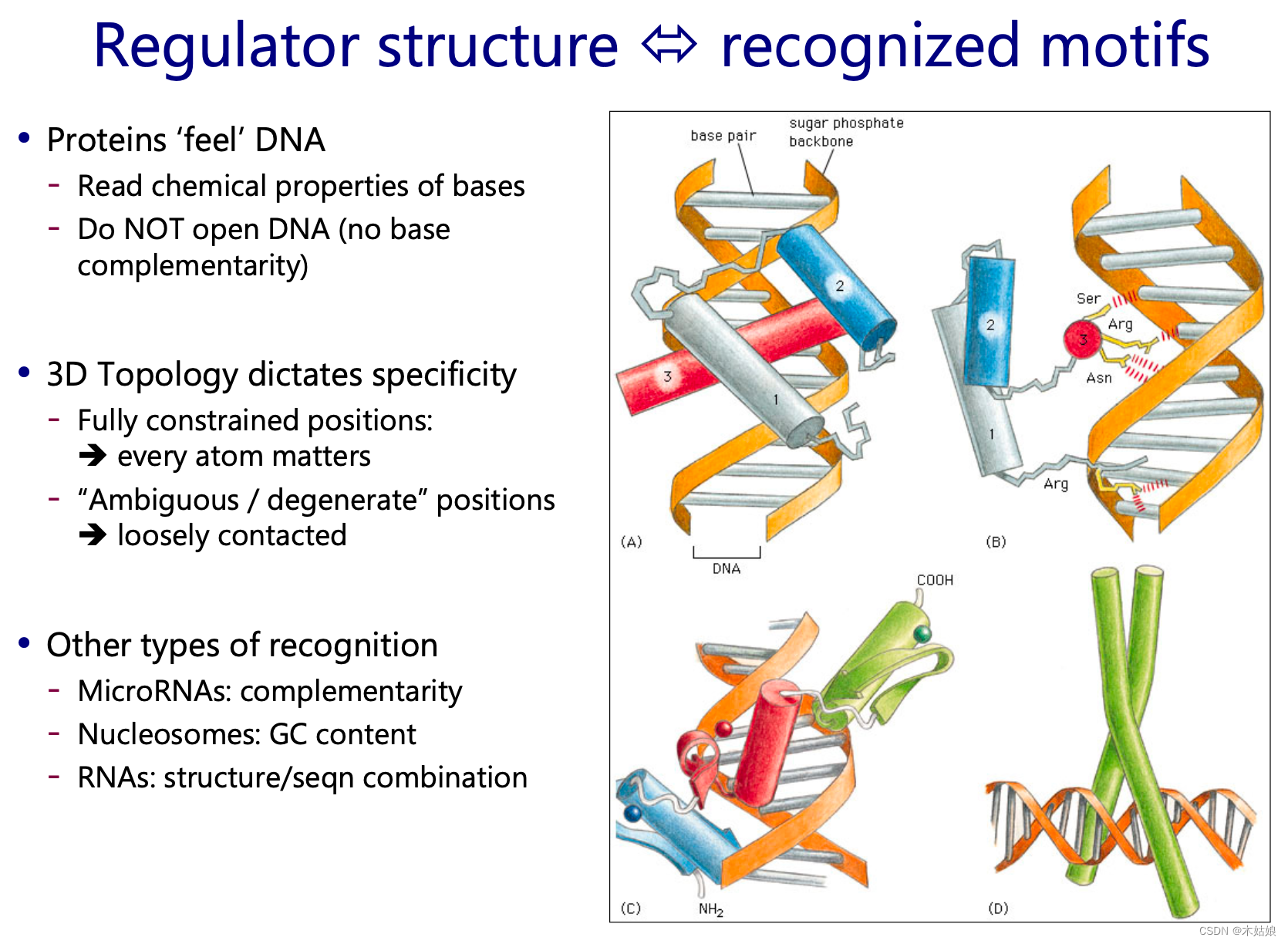
Each protein has a different structure , The binding between proteins is not like DNA,RNA The same complementarity , So they need to go through binding motifs
DNA The representation method of : Pile up all the joints , Then build the matrix , expression DNA The specificity of . The height of the letter indicates the amount of information 


1.2 Probing gene regulation: TFs/histones: ChIP-seq, Accessibility: DNase/ATAC-seq
- DNA Transcription : Histone :ChIP-seq
- DNA accessibility ( Accessibility ): Understand chromatin accessibility and ATAC-seq https://zhuanlan.zhihu.com/p/166500744
2 Traditional methods of regulatory omics and motif discovery
2.1 be based on Enrichment-based Motif discovery :EM Algorithm 、Gibbs Sampling
2.2 An experimental approach :PBMs,SELEX. Comparative genomics:Evolutionary conservation
3 Basic application of convolutional neural network in transcriptional regulation

3.1 Low dimensional features : Main idea :pixels<->DNA letters. Patches/filters<->Motifs. Higher<->combinations
take DNA The sequence is expressed as a two-dimensional matrix 

correct = Ignore signals below certain thresholds .
Pooling = Press max or average Summarize each channel .
Input the extracted features into the network for prediction 
3.2 High dimensional features : Learn convolution kernel <-> Motif discovery. Applying them<-> Motif matches

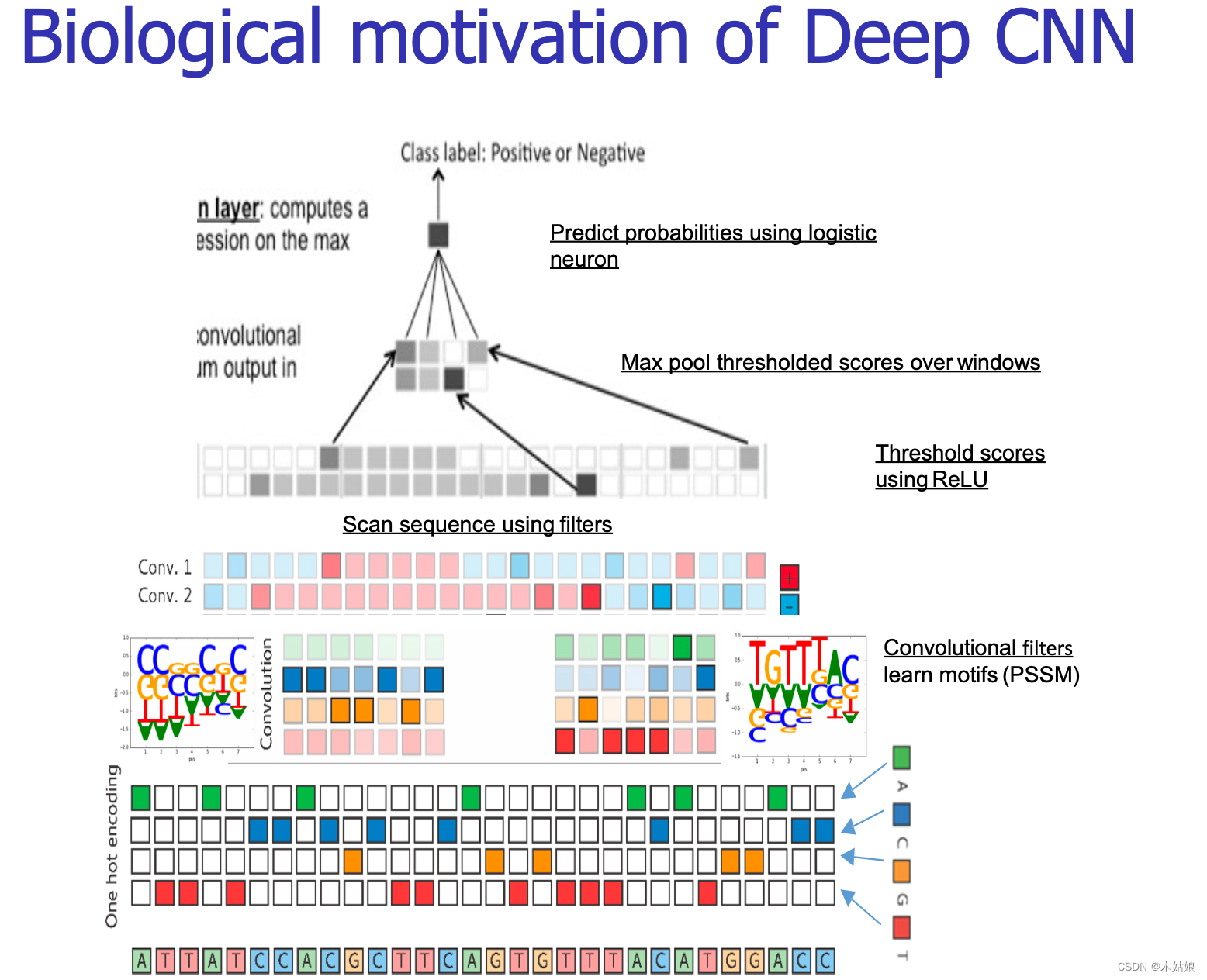
4 CNNS/RNNs Practical application in regulatory omics : Various architectures
4.1 DeepBind: learn motifs, use in(shallow) fully-connected layer, mutations impact
link : http://www.nature.com/nbt/journal/v33/n8/full/nbt.3300.html
DeepBind summary
The key deep learning techniques:
- Convolutional learning
- Representational learning
- Back-propagation and stochastic gradient
- Regularization and dropout
- Parallel GPU computing especially useful for hyper-parameter search
Limitations in DeepBind:
- Require defining negative training examples, which is often arbitrary
- Using observed mutation data only as post-hoc evaluation
- Modeling each regulatory dataset separately
4.2 DeepSea: Train model directly on mutational impact prediction

4.3 Baseet: Multi-task DNase prediction in 164 cell types, reuse/learn motifs
Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks

4.4 ChromPuter: Multi-task prediction of different TFs, reuse partner motifs

4.5 DeepLIFT: Model interpretation based on neuron activation properties

5 Lecture : Stanford Anshul Kundaje,Deep Learning for Reg. Genomics
Deep learning at base-resolution revels cis-regulatory motif syntax
Traditional approach :
Using traditional sequencing methods :ATAC-seq / DNase-seq
Machine learning methods :
Using the accessibility of transcription factors or chromatin , Expression peak, etc. as a label for supervised learning 

BPNet: DNA sequence to base-pair resolution profile regression
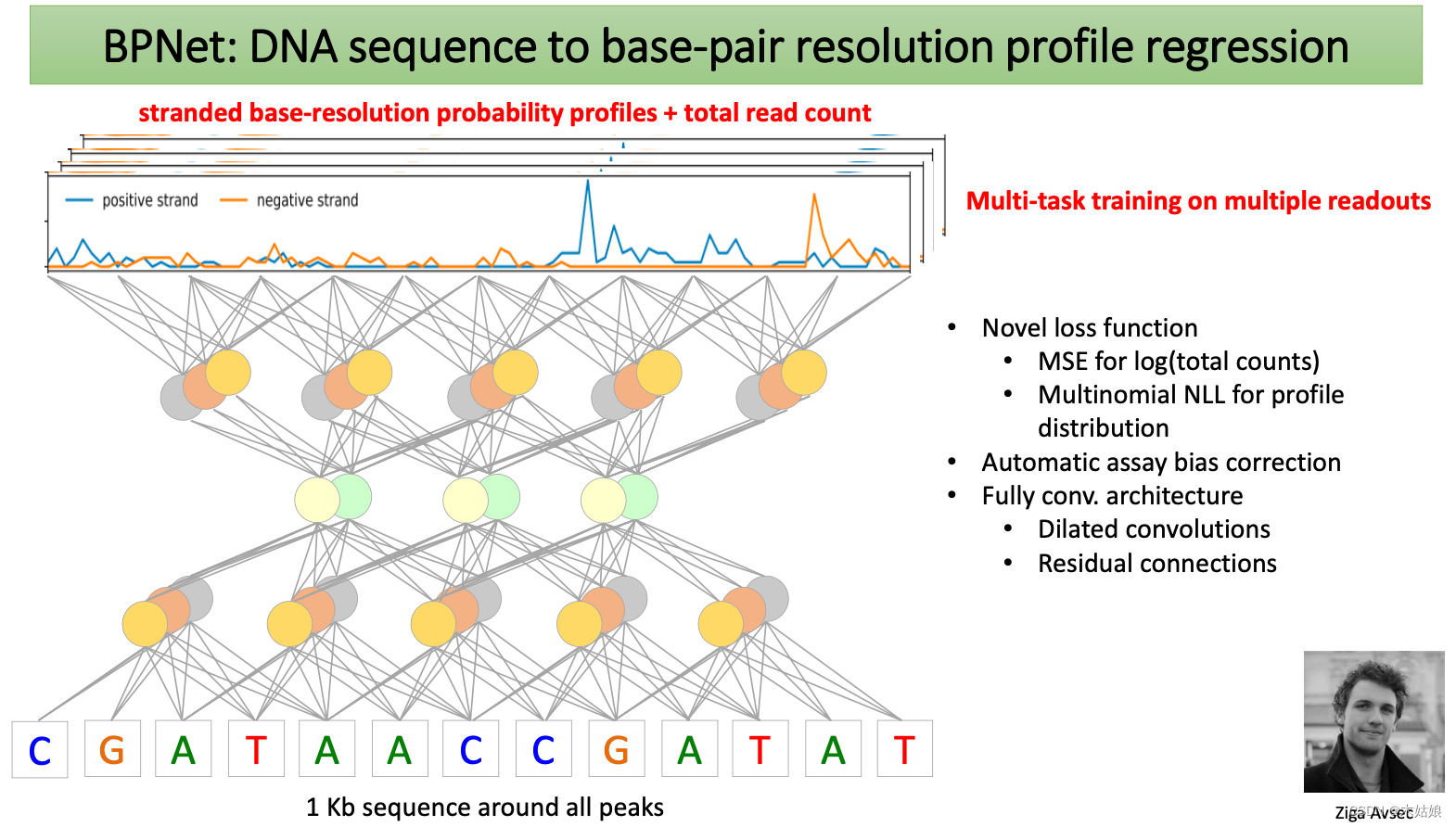
ChIP-exo/nexus: High resolution TF binding footprints

边栏推荐
- 安装和使用
- Appium automation test foundation - ADB shell command
- What makes flutter special
- 栈题目:字符串解码
- [ubuntu-mysql 8 installation and master-slave replication]
- Robot system dynamics - inertia parameters
- keras ‘InputLayer‘ object is not iterable
- Mysql database learning 1
- 1033 To Fill or Not to Fill
- Automated stock trading ensemble strategy based on Reinforcement Learning
猜你喜欢

Detailed explanation of SolidWorks mass characteristics (inertia tensor, moment of inertia, inertia spindle)

How to build a private cloud and create a hybrid cloud ecosystem?

Forrester senior analyst: five important trends in the development of the hyper convergence market

NFS shared services

Rider does not prompt after opening unity script

C語言實現掃雷遊戲,附詳解及完整代碼

力扣 428. 序列化和反序列化 N 叉树 DFS

G code explanation | list of the most important G code commands

机器人系统动力学——惯性参数

【JVM】CMS简述
随机推荐
Follow the wechat oauth2.0 access scheme
[C language quick start] let you know C language and get started with zero basics ③
9. cache optimization
How do databases go to the enterprise cloud? Click to view the answer
《锦绣中华》中老年公益文旅游-走进佛山敬老院
Quick completion guide for manipulator (4): reducer of key components of manipulator
文章内容无法复制复制不了
6.Redis新数据类型
Theme Studio(主题工作室)
JS get the substring of the specified character position and the specified character position interval of the specified string [simple and detailed]
事件对象的说明》
AttributeError: ‘Version‘ object has no attribute ‘major‘
The URL copied by the browser and pasted into the document is a hyperlink
2022第六季完美童模 合肥赛区 初赛圆满落幕
栈题目:字符串解码
MIT-6874-Deep Learning in the Life Sciences Week6
Based on svelte3 X desktop UI component library svelte UI
Regular expression Basics
Galaxy Kirin server-v10 configuration image source
长城数艺数字藏品平台发布创世徽章

