CLASSIX
Fast and explainable clustering based on sorting
CLASSIX is a fast and explainable clustering algorithm based on sorting. Here are a few highlights:
- Ability to cluster low and high-dimensional data of arbitrary shape efficiently.
- Ability to detect and deal with outliers in the data.
- Ability to provide textual explanations for the generated clusters.
- Full reproducibility of all tests in the accompanying paper.
- Support of Cython compilation.
CLASSIX is a contrived acronym of CLustering by Aggregation with Sorting-based Indexing and the letter X for explainability. CLASSIX clustering consists of two phases, namely a greedy aggregation phase of the sorted data into groups of nearby data points, followed by a merging phase of groups into clusters. The algorithm is controlled by two parameters, namely the distance parameter radius for the group aggregation and a minPts parameter controlling the minimal cluster size.
Here is a video abstract of CLASSIX:
A detailed documentation (still in progress), including tutorials, is available at 
Install
CLASSIX has the following dependencies for its clustering functionality:
- cython>=0.29.4
- numpy>=1.20.0
- scipy>1.6.0
- requests
and requires the following packages for data visualization:
- matplotlib
- pandas
To install the current release via PIP use:
pip install ClassixClustering
To install this package with conda run:
conda install -c nla.stefan.xinye classix
To check the installation you can use either of commands (the second is for conda users)
python -m pip show ClassixClustering
conda list classix
Download this repository via:
$ git clone https://github.com/nla-group/classix.git
Quick start
from sklearn import datasets
from classix import CLASSIX
# Generate synthetic data
X, y = datasets.make_blobs(n_samples=1000, centers=2, n_features=2, random_state=1)
# Employ CLASSIX clustering
clx = CLASSIX(sorting='pca', radius=0.5, verbose=0)
clx.fit(X)
Get the clustering result by clx.labels_ and visualize the clustering:
plt.figure(figsize=(10,10))
plt.rcParams['axes.facecolor'] = 'white'
plt.scatter(X[:,0], X[:,1], c=clx.labels_)
plt.show()
The explain method
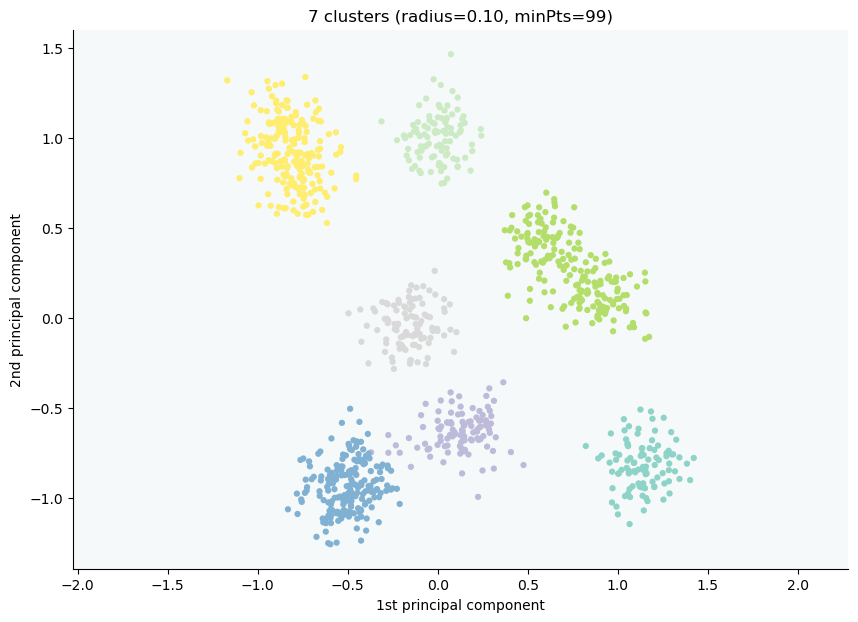
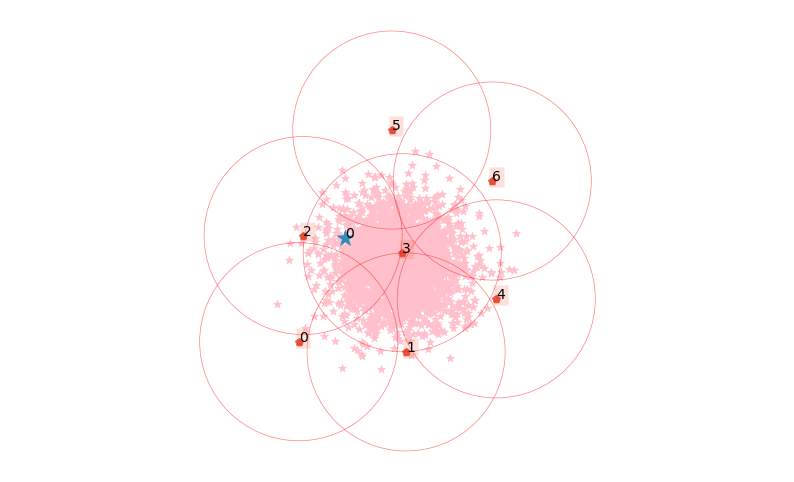
CLASSIX provides an API for the easy visualization of clusters, and to explain the assignment of data points to their clusters. To get an overview of the data points, the location of starting points, and their associated groups, simply type:
clx.explain(plot=True)
The starting points are marked as the small red boxes. The method also returns a textual summary as follows:
A clustering of 5000 data points with 2 features has been performed.
The radius parameter was set to 0.50 and MinPts was set to 0.
As the provided data has been scaled by a factor of 1/6.01,
data points within a radius of R=0.50*6.01=3.01 were aggregated into groups.
In total 7903 comparisons were required (1.58 comparisons per data point).
This resulted in 14 groups, each uniquely associated with a starting point.
These 14 groups were subsequently merged into 2 clusters.
A list of all starting points is shown below.
----------------------------------------
Group NrPts Cluster Coordinates
0 398 0 -1.19 -1.09
1 1073 0 -0.65 -1.15
2 553 0 -1.17 -0.56
--- lines omitted ---
11 6 1 0.42 1.35
12 5 1 1.24 0.59
13 2 1 1.0 1.08
----------------------------------------
In order to explain the clustering of individual data points,
use .explain(ind1) or .explain(ind1, ind2) with indices of the data points.
In the above table, Group denotes the group label, NrPts denotes the number of data points in the group, Cluster is the cluster label assigned to the group, and the final column shows the Coordinates of the starting point. In order to explain the cluster assignment of a particular data point, we provide its index to the explain method:
clx.explain(0, plot=True)
The data point 0 is in group 2, which has been merged into cluster #0.
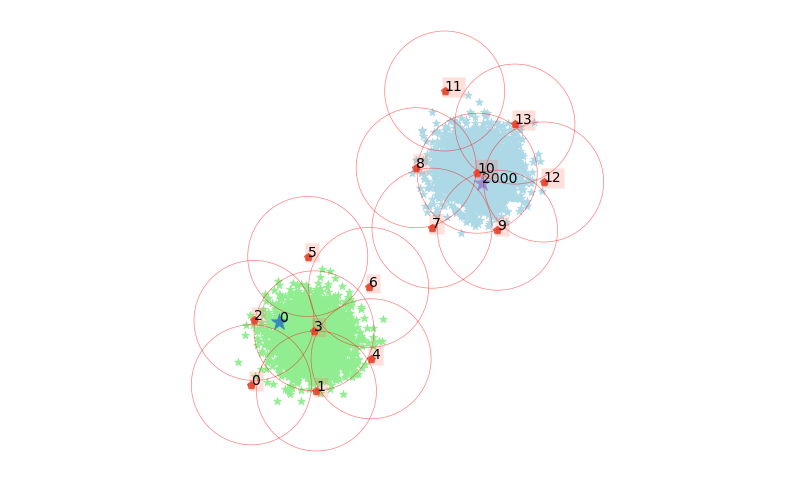
We can also query why two data points ended up in the same cluster, or not:
clx.explain(0, 2000, plot=True)
The data point 0 is in group 2, which has been merged into cluster 0.
The data point 2000 is in group 10, which has been merged into cluster 1.
There is no path of overlapping groups between these clusters.
Reproducible experiment
All empirical data in the paper are reproducible by running the code in the folder of "exp". Before running, ensure the dependency package scikit-learn and hdbscan are installed, and compile Quickshift++ code (obtained in Quickshift++: Provably Good Initializations for Sample-Based Mean Shift). After configuring all of these, run the commands below.
cd exp
python3 run exp_main.py
All results will be stored on "exp/results". Please let us know if you have any questions.
Citation
@techreport{CG22b,
title = {Fast and explainable clustering based on sorting},
author = {Chen, Xinye and G\"{u}ttel, Stefan},
year = {2022},
number = {arXiv:2202.01456},
pages = {25},
institution = {The University of Manchester},
address = {UK},
type = {arXiv EPrint},
url = {https://arxiv.org/abs/2202.01456}
}
License
This project is licensed under the terms of the MIT license.