当前位置:网站首页>Implementation of support vector machine with ml11 sklearn
Implementation of support vector machine with ml11 sklearn
2022-07-29 08:14:00 【19-year-old flower girl】
SKlearn library Realization SVM
%matplotlib inline
# In order to be in notebook Chinese painting display
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
import seaborn as sns; sns.set()
# Random data , Use sklearn Data points are randomly generated by the method under
# among cluster_std Is the degree of dispersion of data
from sklearn.datasets.samples_generator import make_blobs
X, y = make_blobs(n_samples=50, centers=2,
random_state=0, cluster_std=0.60)
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='autumn')

Train a basic SVM
# Classification task
from sklearn.svm import SVC
# Linear kernel function Equivalent to not transforming the data
model = SVC(kernel='linear')
model.fit(X, y)
Template of drawing function
# Plot function
def plot_svc_decision_function(model, ax=None, plot_support=True):
if ax is None:
ax = plt.gca()
xlim = ax.get_xlim()
ylim = ax.get_ylim()
# use SVM Self contained decision_function Function to draw
x = np.linspace(xlim[0], xlim[1], 30)
y = np.linspace(ylim[0], ylim[1], 30)
Y, X = np.meshgrid(y, x)
xy = np.vstack([X.ravel(), Y.ravel()]).T
P = model.decision_function(xy).reshape(X.shape)
# Drawing decision boundaries
ax.contour(X, Y, P, colors='k',
levels=[-1, 0, 1], alpha=0.5,
linestyles=['--', '-', '--'])
# Drawing support vectors
if plot_support:
ax.scatter(model.support_vectors_[:, 0],
model.support_vectors_[:, 1],
s=300, linewidth=1, alpha=0.2);
ax.set_xlim(xlim)
ax.set_ylim(ylim)
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='autumn')
plot_svc_decision_function(model)

This line is the decision boundary we hope to get
It was observed that there was 3 Points are marked with special marks , They happen to be points on the boundary
They are ours support vectors( Support vector )
stay Scikit-Learn in , They are stored in this location support_vectors_( An attribute )
Observation can reveal , We only need support vectors to build the model
Next, let's try , Use different data points , See if the effect will change
Separate use 60 And 120 Data points
def plot_svm(N=10, ax=None):
X, y = make_blobs(n_samples=200, centers=2,
random_state=0, cluster_std=0.60)
X = X[:N]
y = y[:N]
model = SVC(kernel='linear', C=1E10)
model.fit(X, y)
ax = ax or plt.gca()
ax.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='autumn')
ax.set_xlim(-1, 4)
ax.set_ylim(-1, 6)
plot_svc_decision_function(model, ax)
# Draw different data points respectively
fig, ax = plt.subplots(1, 2, figsize=(16, 6))
fig.subplots_adjust(left=0.0625, right=0.95, wspace=0.1)
for axi, N in zip(ax, [60, 120]):
plot_svm(N, axi)
axi.set_title('N = {0}'.format(N))

Introducing kernel functions SVM
Plot another dataset distribution
from sklearn.datasets.samples_generator import make_circles
# Draw another data set
X, y = make_circles(100, factor=.1, noise=.1)
# Let's see if the linear sum function can solve this problem
clf = SVC(kernel='linear').fit(X, y)
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='autumn')
plot_svc_decision_function(clf, plot_support=False);

# Added a new dimension r
from mpl_toolkits import mplot3d
r = np.exp(-(X ** 2).sum(1))
# Imagine stretching a circular dataset up and down in three dimensions
def plot_3D(elev=30, azim=30, X=X, y=y):
ax = plt.subplot(projection='3d')
ax.scatter3D(X[:, 0], X[:, 1], r, c=y, s=50, cmap='autumn')
ax.view_init(elev=elev, azim=azim)
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.set_zlabel('r')
plot_3D(elev=45, azim=45, X=X, y=y)

# Add Gaussian kernel
clf = SVC(kernel='rbf')
clf.fit(X, y)
# This time it's great !
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='autumn')
plot_svc_decision_function(clf)
plt.scatter(clf.support_vectors_[:, 0], clf.support_vectors_[:, 1],
s=300, lw=1, facecolors='none');

Adjust the SVM Parameters

# This data set cluster_std A little bigger , In this way, the function of soft interval can be reflected
X, y = make_blobs(n_samples=100, centers=2,
random_state=0, cluster_std=0.8)
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='autumn')

C Parameters
# Data sets that make the game more difficult
X, y = make_blobs(n_samples=100, centers=2,
random_state=0, cluster_std=0.8)
fig, ax = plt.subplots(1, 2, figsize=(16, 6))
fig.subplots_adjust(left=0.0625, right=0.95, wspace=0.1)
# Select two C Parameters to carry out the opposite experiment , Respectively 10 and 0.1
for axi, C in zip(ax, [10.0, 0.1]):
model = SVC(kernel='linear', C=C).fit(X, y)
axi.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='autumn')
plot_svc_decision_function(model, axi)
axi.scatter(model.support_vectors_[:, 0],
model.support_vectors_[:, 1],
s=300, lw=1, facecolors='none');
axi.set_title('C = {0:.1f}'.format(C), size=14)

Karma parameter , The larger the mapping, the higher the dimension , The more complex the model .
X, y = make_blobs(n_samples=100, centers=2,
random_state=0, cluster_std=1.1)
fig, ax = plt.subplots(1, 2, figsize=(16, 6))
fig.subplots_adjust(left=0.0625, right=0.95, wspace=0.1)
# Choose different gamma Value to observe the modeling effect
for axi, gamma in zip(ax, [10.0, 0.1]):
model = SVC(kernel='rbf', gamma=gamma).fit(X, y)
axi.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='autumn')
plot_svc_decision_function(model, axi)
axi.scatter(model.support_vectors_[:, 0],
model.support_vectors_[:, 1],
s=300, lw=1, facecolors='none');
axi.set_title('gamma = {0:.1f}'.format(gamma), size=14)
For relatively large karma value , The boundary is clearly divided , But the generalization ability is relatively low ; Smaller karma value , Wrong data points , But the generalization ability is strong , More useful .
Face recognition examples
# Reading data sets
from sklearn.datasets import fetch_lfw_people
# Everyone's face has at least 60 individual
faces = fetch_lfw_people(min_faces_per_person=60)
# Look at the scale of the data
print(faces.target_names)
print(faces.images.shape)
# 3 That's ok 5 Column layout
fig, ax = plt.subplots(3, 5)
for i, axi in enumerate(ax.flat):
axi.imshow(faces.images[i], cmap='bone')
axi.set(xticks=[], yticks=[],
xlabel=faces.target_names[faces.target[i]])


from sklearn.svm import SVC
from sklearn.decomposition import PCA
from sklearn.pipeline import make_pipeline
# Dimensionality reduction 150 dimension
pca = PCA(n_components=150, whiten=True, random_state=42)
svc = SVC(kernel='rbf', class_weight='balanced')
# First reduce the dimension and then SVM
model = make_pipeline(pca, svc)
Divide the data set
from sklearn.model_selection import train_test_split
Xtrain, Xtest, ytrain, ytest = train_test_split(faces.data, faces.target,
random_state=40)
Use grid search cross-validation To choose our parameters , Traverse C Hegama , See which works well .
from sklearn.model_selection import GridSearchCV
param_grid = {
'svc__C': [1, 5, 10],
'svc__gamma': [0.0001, 0.0005, 0.001]}
grid = GridSearchCV(model, param_grid)
%time grid.fit(Xtrain, ytrain)
print(grid.best_params_)
After selection, use our model to make predictions .
model = grid.best_estimator_
yfit = model.predict(Xtest)
yfit.shape
Result display
fig, ax = plt.subplots(4, 6)
for i, axi in enumerate(ax.flat):
axi.imshow(Xtest[i].reshape(62, 47), cmap='bone')
axi.set(xticks=[], yticks=[])
axi.set_ylabel(faces.target_names[yfit[i]].split()[-1],
color='black' if yfit[i] == ytest[i] else 'red')
fig.suptitle('Predicted Names; Incorrect Labels in Red', size=14);

from sklearn.metrics import classification_report
print(classification_report(ytest, yfit,
target_names=faces.target_names))
Precision value and recall rate 
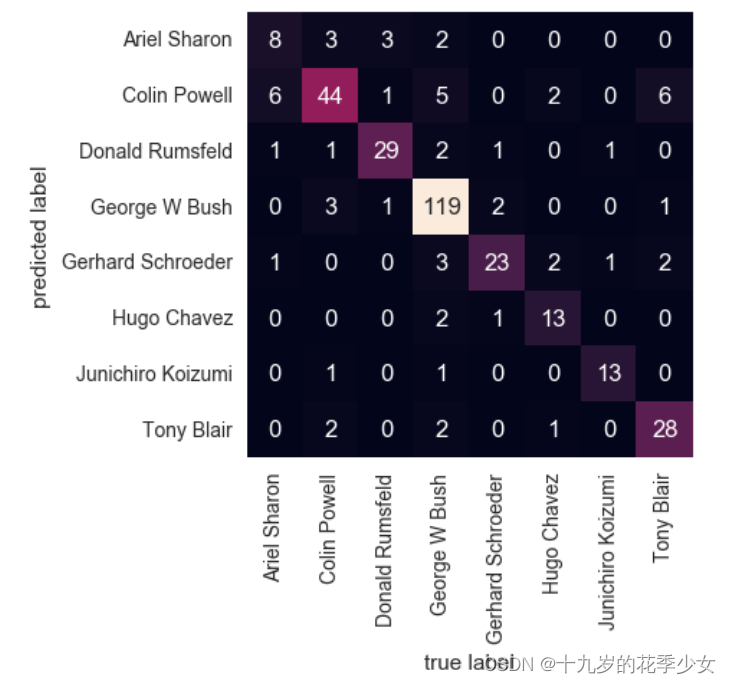
Confusion matrix
from sklearn.metrics import confusion_matrix
mat = confusion_matrix(ytest, yfit)
sns.heatmap(mat.T, square=True, annot=True, fmt='d', cbar=False,
xticklabels=faces.target_names,
yticklabels=faces.target_names)
plt.xlabel('true label')
plt.ylabel('predicted label');
边栏推荐
- File system I
- NFC two-way communication 13.56MHz contactless reader chip -- si512 replaces pn512
- Dp1332e multi protocol highly integrated contactless read-write chip
- Crawl notes
- The computer system has no standard tcp/ip port processing operations
- Inclination sensor accuracy calibration test
- Very practical shell and shellcheck
- Privacy is more secure in the era of digital RMB
- An optimal buffer management scheme with dynamic thresholds paper summary
- Crawl expression bag
猜你喜欢

Solve the problem of MSVC2017 compiler with yellow exclamation mark in kits component of QT

Windows 安装 MySQL 5.7详细步骤

Alibaba political commissar system - Chapter 4: political commissars are built on companies

Unicode私人使用区域(Private Use Areas)

Ansible (automation software)

Cs5340 domestic alternative dp5340 multi bit audio a/d converter

How to connect VMware virtual machine to external network under physical machine win10 system

Official tutorial redshift 01 basic theoretical knowledge and basic characteristics learning

Cv520 domestic replacement of ci521 13.56MHz contactless reader chip
![[beauty of software engineering - column notes] 23 | Architect: programmers who don't want to be architects are not good programmers](/img/a2/020da8a88e7c68f3dcca48208baff2.png)
[beauty of software engineering - column notes] 23 | Architect: programmers who don't want to be architects are not good programmers
随机推荐
Network Security Learning chapter
Tle5012b+stm32f103c8t6 (bluepill) reading angle data
Operator overloading
Proteus simulation based on msp430f2491
Low power Bluetooth 5.0 chip nrf52832-qfaa
torch.Tensor和torch.tensor的区别
Solve the problem that the disk is full due to large files
20 hacker artifacts
[cryptography experiment] 0x00 install NTL Library
torch.nn.functional.one_ hot()
Simple calculator wechat applet project source code
UE4 highlight official reference value
DC motor control system based on DAC0832
Shell script - global variables, local variables, environment variables
Cv520 domestic replacement of ci521 13.56MHz contactless reader chip
阿里巴巴政委体系-第一章、政委建在连队上
Processes and threads
Tcp/ip five layer reference model and corresponding typical devices and IPv6
Day 014 2D array exercise
Unity beginner 1 - character movement control (2D)