pmdarima
Pmdarima (originally pyramid-arima, for the anagram of 'py' + 'arima') is a statistical library designed to fill the void in Python's time series analysis capabilities. This includes:
- The equivalent of R's
auto.arimafunctionality - A collection of statistical tests of stationarity and seasonality
- Time series utilities, such as differencing and inverse differencing
- Numerous endogenous and exogenous transformers and featurizers, including Box-Cox and Fourier transformations
- Seasonal time series decompositions
- Cross-validation utilities
- A rich collection of built-in time series datasets for prototyping and examples
- Scikit-learn-esque pipelines to consolidate your estimators and promote productionization
Pmdarima wraps statsmodels under the hood, but is designed with an interface that's familiar to users coming from a scikit-learn background.
Installation
pip
Pmdarima has binary and source distributions for Windows, Mac and Linux (manylinux) on pypi under the package name pmdarima and can be downloaded via pip:
pip install pmdarima
conda
Pmdarima also has Mac and Linux builds available via conda and can be installed like so:
conda config --add channels conda-forge
conda config --set channel_priority strict
conda install pmdarima
Note: We do not maintain our own Conda binaries, they are maintained at https://github.com/conda-forge/pmdarima-feedstock. See that repo for further documentation on working with Pmdarima on Conda.
Quickstart Examples
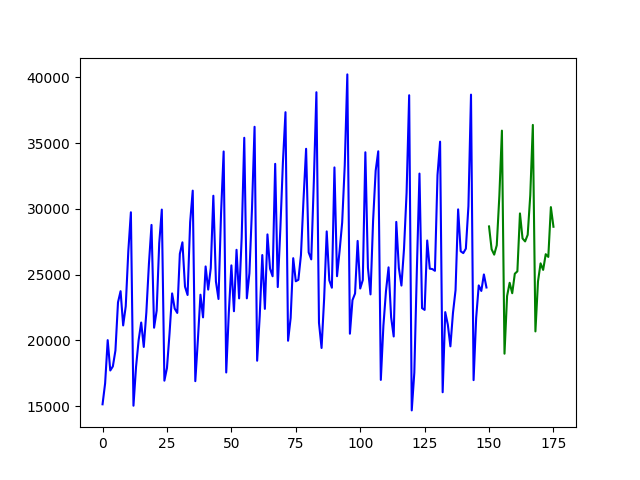
Fitting a simple auto-ARIMA on the wineind dataset:
import pmdarima as pm
from pmdarima.model_selection import train_test_split
import numpy as np
import matplotlib.pyplot as plt
# Load/split your data
y = pm.datasets.load_wineind()
train, test = train_test_split(y, train_size=150)
# Fit your model
model = pm.auto_arima(train, seasonal=True, m=12)
# make your forecasts
forecasts = model.predict(test.shape[0]) # predict N steps into the future
# Visualize the forecasts (blue=train, green=forecasts)
x = np.arange(y.shape[0])
plt.plot(x[:150], train, c='blue')
plt.plot(x[150:], forecasts, c='green')
plt.show()
Fitting a more complex pipeline on the sunspots dataset, serializing it, and then loading it from disk to make predictions:
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
Availability
pmdarima is available on PyPi in pre-built Wheel files for Python 3.6+ for the following platforms:
- Mac (64-bit)
- Linux (64-bit manylinux)
- Windows (32 & 64-bit)
If a wheel doesn't exist for your platform, you can still pip install and it will build from the source distribution tarball, however you'll need cython>=0.29 and gcc (Mac/Linux) or MinGW (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name "pyramid-arima" and can be pip installed via:
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
However, this is not recommended.
Documentation
All of your questions and more (including examples and guides) can be answered by the pmdarima documentation. If not, always feel free to file an issue.