当前位置:网站首页>[advanced drawing of single cell] 07. Display of KEGG enrichment results
[advanced drawing of single cell] 07. Display of KEGG enrichment results
2022-07-28 08:59:00 【Top bioinformation】
 The pictures drawn in this section are relatively new , What I marked with red arrows in the picture is pathway
The pictures drawn in this section are relatively new , What I marked with red arrows in the picture is pathway Class A Annotation information (big annotation, What I think , It's not a proper noun ), The colorful marks on the vertical axis are pathway Of second level notes (small annotation).「 How to get comments 」 Calculate a difficult point , I Last time I've already said :KEGG The subordination of the path / How to get annotation information .
The whole picture reflects How many genes Fell into the corresponding classification .
「 Look at it dialectically 」, The whole picture is pathway notes , There is no specific pathway name , It is very similar to the usual enrichment analysis Dissimilarity . Replace the secondary notes in the figure with specific pathway Will be better . in addition , The abscissa in this figure is Gene number , But I don't think this value is important in enrichment analysis ( gene ratio More important than a single value ), We can change it to p value .
Before you start drawing , Need to sort out a form , The table contains at least :「pathway ID、pathway description、pathway notes /big annotation、 Several enrichment indicators 、 Enriched genes 」.
「 The first three messages 」, The forms of the previous lecture have been sorted out , It can be used ; 「 Several enrichment indicators 」 stay clusterprofilerThere are also ;「 Enriched genes 」 Looks like clusterprofiler do kegg Enrichment cannot directly show genes symbol, So the returned result needs to be processed a little .
The code name of the sorting table is run.R, As shown below :
library(Seurat)
library(tidyverse)
library(xlsx)
testseu=readRDS("testseu.rds")
Idents(testseu)="anno_new"
### Look for differential genes #########################################################################
marker_celltype=FindAllMarkers(testseu,logfc.threshold = 0.8,only.pos = T)
# Filter
marker_celltype=marker_celltype%>%filter(p_val_adj < 0.01)
marker_celltype$d=marker_celltype$pct.1-marker_celltype$pct.2
marker_celltype=marker_celltype%>%filter(d > 0.2)
marker_celltype=marker_celltype%>%arrange(cluster,desc(avg_log2FC))
marker_celltype=as.data.frame(marker_celltype)
write.xlsx(marker_celltype,file = "markers_log2fc0.8_padj0.01_pctd0.2.xlsx",row.names = F,col.names = T)
### Enrichment analysis ###########################################################################
library(clusterProfiler)
library(org.Hs.eg.db)
R.utils::setOption("clusterProfiler.download.method","auto") #https://github.com/YuLab-SMU/clusterProfiler/issues/256
source("syEnrich.R")
syEnrich(marker_celltype,outpath = "markers_log2fc0.8_padj0.01_pctd0.2")
### Select a type of cell as a demonstration #######################################################
kegg.res=read.xlsx("markers_log2fc0.8_padj0.01_pctd0.2.KEGG.xls",sheetIndex = 1,as.data.frame = T,header = T)
kegg.res=kegg.res%>%filter(p.adjust < 0.05)
kegg.res=kegg.res%>%filter(cluster == "Endothelial")
# Import the file mentioned above
kegg_info=read.xlsx("kegg_info.xlsx",sheetIndex = 1,startRow = 3)
kegg_info=kegg_info[,c("ID","Pathway","big.annotion")]
# Merge two tables
kegg.res$ID=str_replace(kegg.res$ID,"hsa","")
kegg.res=kegg.res%>%inner_join(kegg_info,by = "ID")
write.table(kegg.res,file = "kegg.res.txt",quote = F,sep = "\t",row.names = F,col.names = T)
I take Single cell analysis Medium kegg Enrichment analysis as a demonstration , Just take one of them cluster Draw a picture based on the enrichment results of .
The enrichment code used in the above code is called syEnrich.R, This file only needs to input single cells seurat Object running FindAllMarkers The resulting differential genes , I can go back GO/KEGG Enrichment results , At the same time, genes enriched into a certain pathway symbol Will also be listed .
function run.R after , The final table is shown in the figure below : 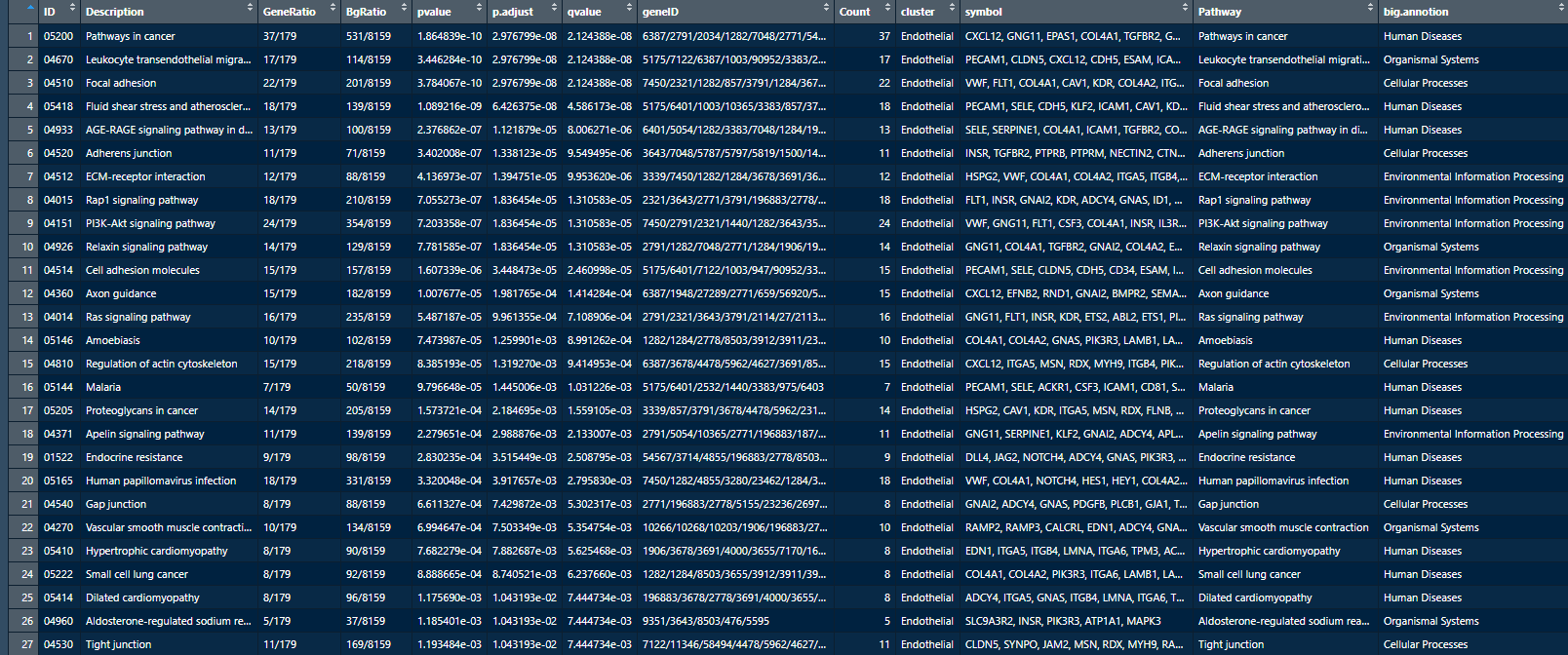
Then start drawing , The code name is 3.plot.R, I'm not going to do that here , The final figure is as follows :
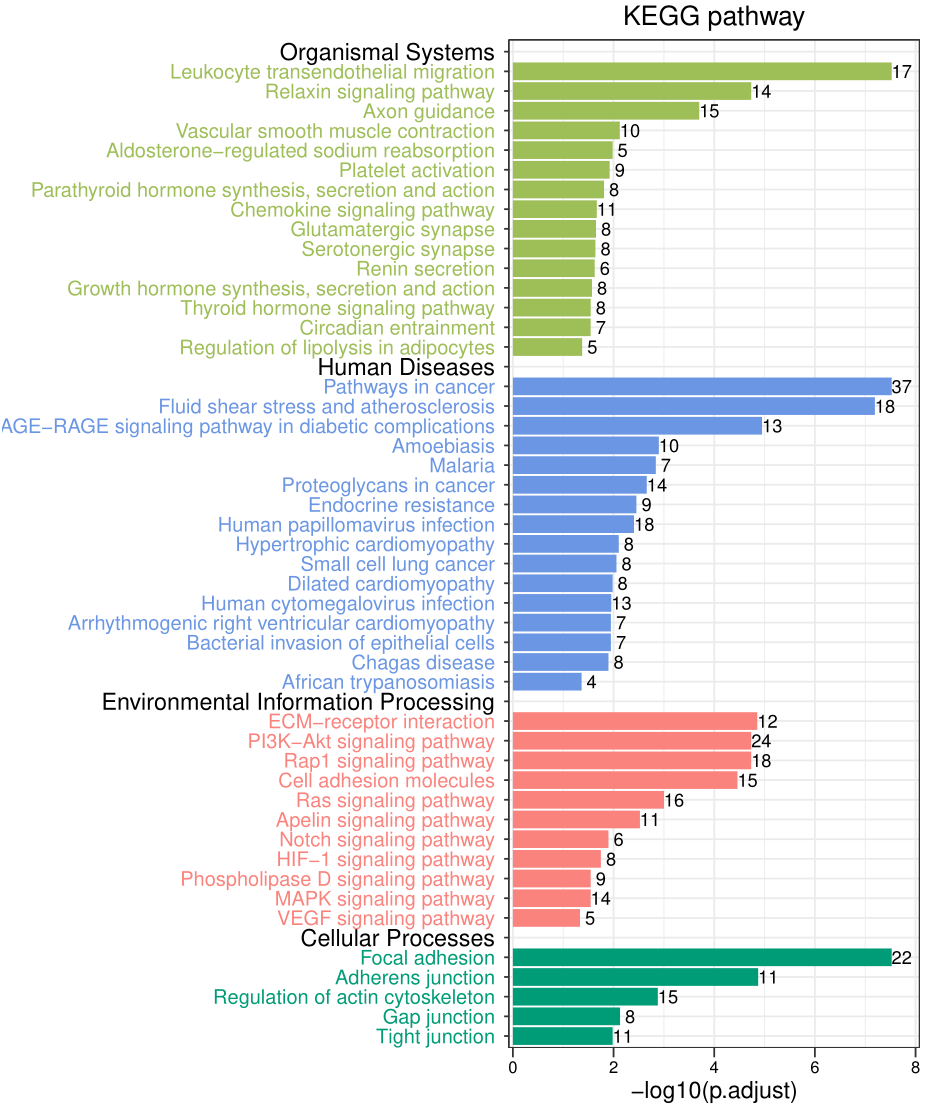
Get the code
Include all the code that will be used in this figure , Data sorting and drawing , Is it super sweet !
This series will take 「 Time limited disclosure 」 Share code in a way ,24 Hours The inside is free . How to get beyond this time , The background to reply 2022A You know ( You may need to use your little hand to forward ).
The network disk link of code and test data is as follows : link :https://pan.baidu.com/s/1hebbeQH4DgYA8JqFWXO4dg Extraction code :abo5
If you think the code is useful , You can give three / Two / One after another ( At the end of the article give the thumbs-up 、 Share 、 Click the small advertisement ) Help me :( A few times ago, there was a pitiful rise
边栏推荐
- Three ways to create threads
- 分布式系统架构理论与组件
- Business digitalization is running rapidly, and management digitalization urgently needs to start
- Why setting application.targetframerate doesn't work
- 第2章-14 求整数段和
- Recruiting talents, gbase high-end talent recruitment in progress
- 创建线程的3种方式
- A new method of exposing services in kubernetes clusters
- [opencv] generate transparent PNG image
- 【单细胞高级绘图】07.KEGG富集结果展示
猜你喜欢
![[opencv] generate transparent PNG image](/img/0a/4afc9bda411634562f4b0f3915a7ba.png)
[opencv] generate transparent PNG image

Among China's top ten national snacks, it is actually the first

Bluetooth technology | it is reported that apple, meta and other manufacturers will promote new wearable devices, and Bluetooth will help the development of intelligent wearable devices

Go waitgroup and defer

1w5 words to introduce those technical solutions of distributed system in detail

Hcip day 9_ BGP experiment

After reading these 12 interview questions, the new media operation post is yours

JS inheritance method

ciou损失

Solution: indexerror: index 13 is out of bounds for dimension 0 with size 13
随机推荐
Introduction to self drive tour of snow mountains in the West in January 2018
SQL injection - pre Foundation
[activity registration] User Group Xi'an - empowering enterprise growth with modern data architecture
Vrrp+mstp configuration details [Huawei ENSP experiment]
[cloud computing] several mistakes that enterprises need to avoid after going to the cloud
我来教你如何组装一个注册中心?
Larkapi access credentials overview
This flick SQL timestamp_ Can ltz be used in create DDL
How to configure phpunit under window
PostgreSQL:无法更改视图或规则使用的列的类型
1299_ Task status and switching test in FreeRTOS
Shell programming specifications and variables
Two dimensional array and operation
为什么 ThreadLocal 可以做到线程隔离?
I am a 27 year old technical manager, whose income is too high, and my heart is in a panic
Div tags and span Tags
In addition to exporting the incremental data captured by Oracle golden gate to Oracle, where can it be exported? Can be similar
Why is the text box of Google material design not used?
A new method of exposing services in kubernetes clusters
Hcip day 9_ BGP experiment