当前位置:网站首页>Nature microbiology | viral genomes in six deep-sea sediments that can infect Archaea asgardii
Nature microbiology | viral genomes in six deep-sea sediments that can infect Archaea asgardii
2022-07-04 18:40:00 【Liu Yongxin Adam】

translate : Zhou Zhichao @UW-Madison
Genomes of six viruses that infect Asgard archaea from deep-sea sediments
DOI:10.1038/s41564-022-01150-8
Link to the original text :https://doi.org/10.1038/s41564-022-01150-8
First author :Ian M. Rambo
Corresponding author :Brett J. Baker
Main units :Department of Marine Science, University of Texas Austin( Department of marine science, University of Texas at Austin )
Abstract
Archaea asgardii is a worldwide distribution of prokaryotic microorganisms related to eukaryotes ; However , The viruses that infect these organisms have not been described . ad locum , Using macrogenomic sequences extracted from deep-sea hydrothermal sediments , We described 6 Relatively large ( reach 117 kb) Double chain DNA(dsDNA) Viral genome , They are infected with two archaeopteroides ,Lokiarchaeota and Helarchaeota. These viruses encode something similar to Caudovirales Structural proteins , And proteins different from those described in known paleontological viruses . Their genomes contain large amounts of eukaryotic cytoplasm DNA Viruses (NCLDVs) Related about 1-5% Genes , It seems that semi autonomous genome replication can be carried out 、 Repair 、 Epigenetic modification and transcriptional regulation . Besides ,Helarchaeota The virus may hijack the host's ubiquitin system , Similar to eukaryotic viruses . Genome analysis of these Asgard viruses revealed , They contain both prokaryotic and eukaryotic virus characteristics , This study provides new insights into the mechanism of potential infection and host interaction .
Text
Archaea asgardii is a globally distributed microorganism , Related to eukaryotes . Their genome composition shows that they are the descendants of the Archaea host that produced the first common ancestor of eukaryotes . In recent years , Due to the recovery of the genome from a series of marine and terrestrial aquatic sediments , Asgard's biodiversity has greatly expanded . An anaerobic 、 Slow growing Asgard pedigree ,Lokiarchaeota, Recently cultivated , It seems to depend on bacteria for mutual nourishment . This supports a variety of genome-based Asgard - Inference of bacterial interaction , These inferences are thought to have led to the formation of the first eukaryotic cell containing mitochondria . Others assume , The interaction with viruses contributed to the origin of complex cell life . This is based on some NCLDVs And the presence of bacteriophage nucleovirus factories , It allows replication in the host cytoplasm , And pass mRNA Capping decouples transcription from Translation . stay Lokiarchaeota The putative viral protein has been found in the genome of , This shows that the virus played a role in the exchange of genetic elements and the evolution of Archaea asgardii .I The type and III type CRISPR-Cas The immune system has been in several Asgard gates (Odinarchaeota、Thorarchaeota、Lokiarchaeota and Helarchaeota) Described in , But so far, the viral genome related to Archaea asgardii has not been characterized .
To explore the role of viruses in archaea asgardii , We searched for Guaymas basin (~2,000 Meters deep , The Gulf of California ) In hydrothermal related sediments ~5TB Metagenome sequence data (assemblies). from , We recovered 6,756 Uncultured viral genome (UViGs)( chart 1), It is estimated to be of high quality and medium quality ( See method ).UViGs And from Guaymas Basin metagenome assembles genome (MAGs) Of CRISPR Interval sequence (spacer) Connected to a , To determine which viruses infected Archaea asgardii cells . This reveals 7 A double chain DNA(dsDNA)UViG, They are infected with Lokiarchaeota、Helarchaeota and Thorarchaeota( chart 2). Interestingly , Two viruses related to Archaea asgardii have also been infected from Guaymas Basin reconstruction Chloroflexi and Omnitrophica bacteria , It shows that Asgard is closely related to these bacteria . One of them UViG yes Chloroflexi Integrated protoviruses in the genome , And Thorarchaeota Yes CRISPR Interval sequence connection , The other six are non integrated , It is classified as cracking type . Let's name these creatures in Nordic mythology UViG.“Vedfolnir'”(Chloroflexi provirus And Thorarchaeota Related to )、Fenrir(Lokiarchaeota)、Sköll(Lokiarchaeota)、Nidhogg(Helarchaeota) as well as Ratatoskr(Helarchaeota and Omnitrophica). Their genome size ( about 21.9-117.5kb) Belonging to known Archaea dsDNA The scope of the virus ( chart 2). from Meg22_1012 Found in sample No Nidhogg and Fenrir Viruses and Meg22_1214 The nucleotide consistency of sample No. is very high ( Respectively 99.98 and 99.90%).Nidhogg The virus is enclosed into a complete circular genome ( chart 2D), Two linear Fenrir The virus is predicted to be >90% The integrity of .
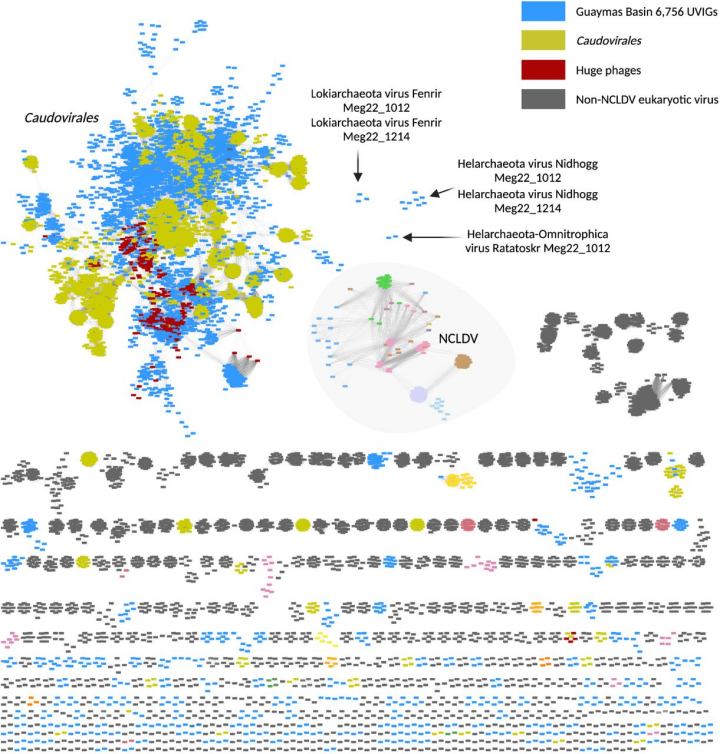
chart 1. from Guaymas The protein clustering network of the viral genome found in the sediments of the basin compared with the previously described viruses .
use vContact2 v.0.9.19 and Cytoscape v.3.8.0 Built monomer network analysis , Yes Guaymas Of the basin UViG(n = 6,756, Displayed in light blue ) Classify and assign , Use from RefSeq Reference eukaryotes 、 Bacterial and archaeovirus genomes (n = 11,082) And giant bacteriophages (n = 361)( See method ). Nodes represent a single genome , The edge represents the similarity between genomes within the virus cluster (n = 770). The virus associated with Archaea asgardii is labeled Fenrir、Nidhogg and Ratatoskr.Vedfolnir and Sköll Be classified as outliers (outliers).
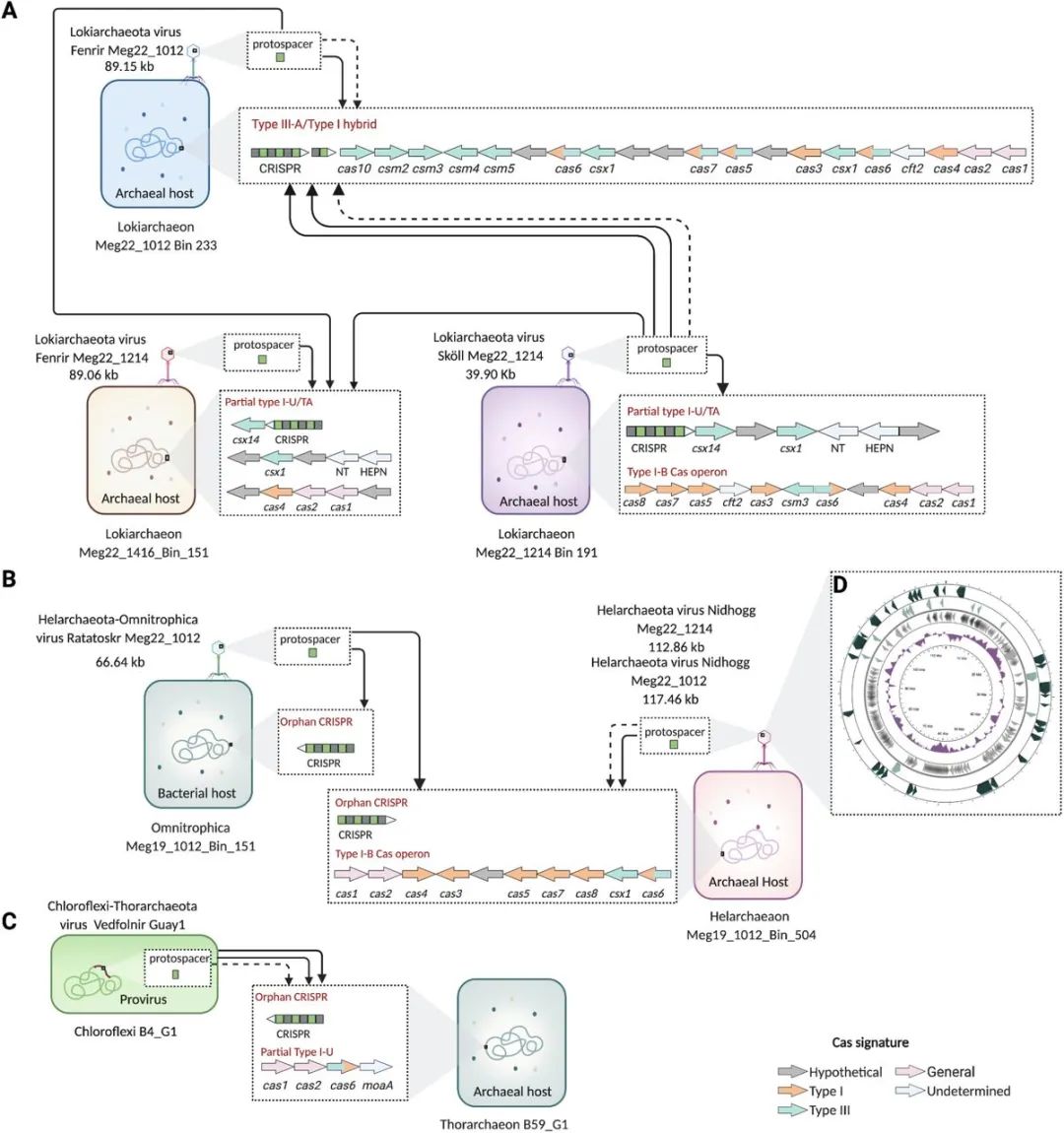
chart 2. Viruses - The connection between the host and archaea asgardii .
(A)Lokiarchaeota, (B)Helarchaeota, and (C)Thorarchaeota MAGs adopt CRISPR-Cas System and UViG Connected to a .(D) Complete ring Nidhogg Overview of viral genome .CRISPR Spacer bar and UViG Of blastn-short alignments Show , The arrow stands for 100% The consistency of ( Solid line ) and 95%-99.9% The consistency of ( Dotted line ). In this study Lokiarchaeaota、Helarchaeota and Omnitrophica MAGs.Chloroflexi and Thorarchaeota The genome of has been previously restored . Predicted Asgard MAGs Of CRISPR-Cas The system is indicated by an arrow , According to its Cas The type characteristics of genes are colored , The arrow pointing to the right is (+), Justice , On the left (-) That is, antisense .CRISPR The arrows at the end of the array represent justice or antisense in the same way . Each... Displayed cas The boxes are located in different scaffold On . Viruses are represented by bacteriophage like icons , Even though there is only Ratatoskr A shell containing a recognizable icosahedron .Nidhogg The coding region in the genome points to the right (+), On the left (-), And draw to scale .cas1,CRISPR Related endonuclease Cas1;cas2,CRISPR Related endonuclease Cas2;cas3,CRISPR Related endonuclease / Helicase Cas3;cas4,CRISPR Related exoenzymes Cas4;cas5,CRISPR System Cascade Subunits Cas5.cas6,CRISPR Related endonuclease Cas6;cas7,CRISPR-Cas The first type of effector complex subunit Cas7;cas8,CRISPR Related proteins Cas8;csm2, The third category CSM Small subunit of effector complex Csm2;csm3, The third category RAMP Superfamily CSM Effector complex Csm3.csm4, III type RAMP Superfamily CSM Effector complex Csm4; csm5, III type RAMP Superfamily CSM Effector complex Csm5; csx1, CRISPR System endonuclease Csx1; csx14, III-U Subtype related proteins Csx14.cft2,Cft2 family RNA Deal with exonuclease ;MoaA, Molybdenum cofactor biosynthetic protein MoaA;TA, Toxins - Antitoxin ;NT, Nucleotide transferase ;HEPN, Nucleotide binding domains in higher eukaryotes and prokaryotes ;kb, Kilobase . use BioRender.com establish .
In order to classify and understand the evolutionary history of Asgard virus , We identified seven UViG Medium DNA Polymerase B(PolB) protein . come from Fenrir、Nidhogg and Ratatoskr Of PolB Protein sequences and derived from cellular organisms 、 Phages 、 Archaea and eukaryotic viruses and NCLDVs Of PolB Compared with . Phylogenetic analysis of these sequences revealed ,Fenrir、Nidhogg and Ratatoskr Different from previous characteristic viruses , With a complex evolutionary history ( chart 3A).Fenrir PolB And Gammaproteobacteria The close relationship between , These genes are transferred horizontally from bacteria , and Nidhogg PolB It seems that the host is Halobacillus myoviruses There is a phylogenetic relationship ( chart 3A).Ratatoskr PolB Phylogeny with Heimdallarchaeota of ,Heimdallarchaeota It is considered to have the closest relationship with eukaryotes Asgard door ( chart 3B). In order to determine the family level classification of Asgard virus , We study its viral protein family (VPF) The composition of . All seven Asgard UViGs Both contain and Myoviridae、Siphoviridae and Podoviridae As well as NCLDVs Gene homologues related to thermophilic archaiviridae ( chart 3C).Asgard UViGs Do not cluster with existing reference viruses based on unilateral gene sharing networks ( chart 1), This may indicate that they are Caudovirales A new family or genus of . This is supported by previous studies , These studies identify global Caudovirales And archaea . There is no clear subordinate relationship with the previously described virus , It highlights the novelty of these newly discovered Asgard viruses .
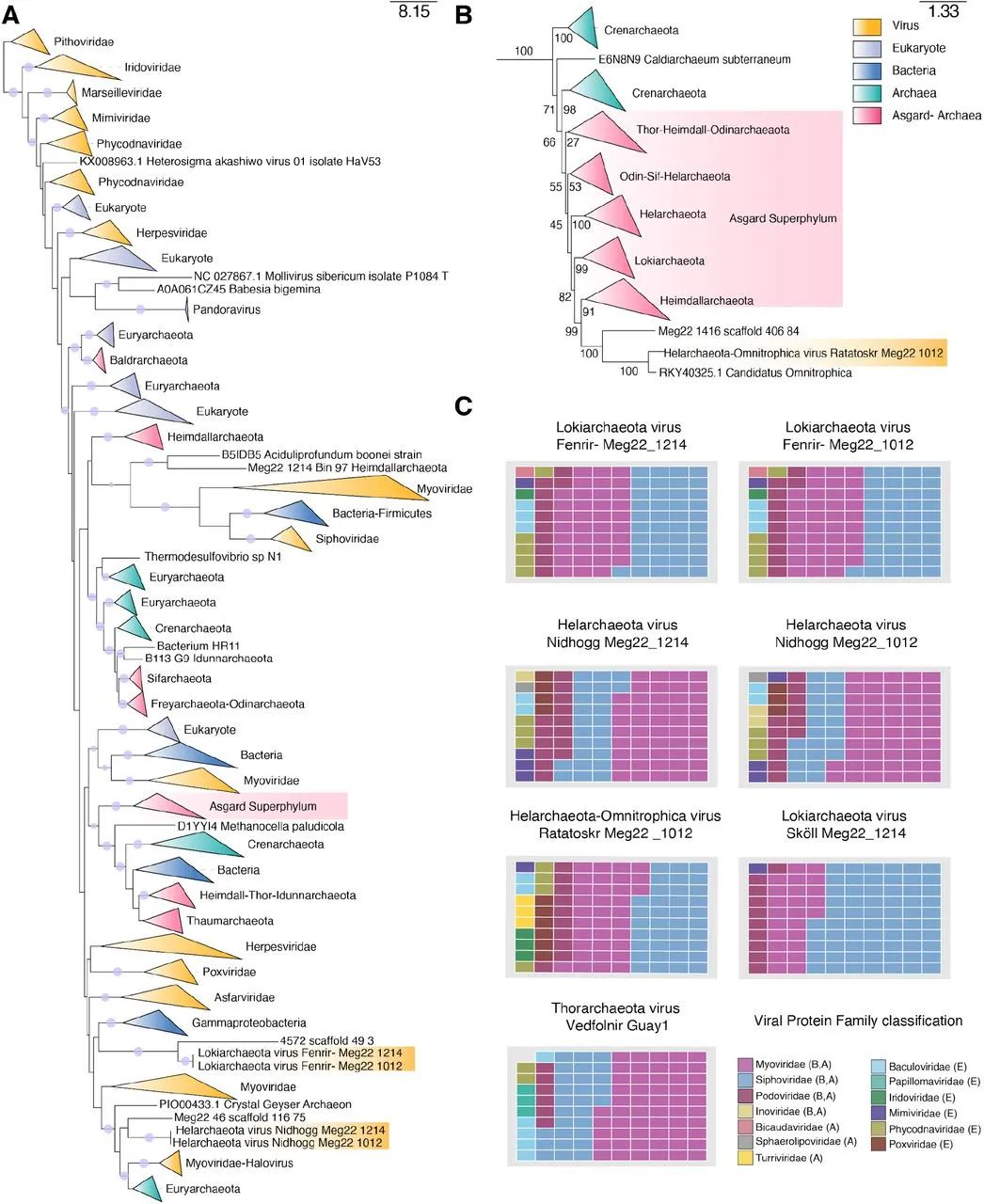
chart 3. The taxonomic location of Asgard virus based on phylogeny and protein composition .
(A) Use LG+F+R10 Model generated DNA Polymerase B Maximum likelihood phylogenetic tree . The circles on the branches represent ultrafast bootstrap rate of support >=95. The viruses described in this article are highlighted in gold .(B) Ratatoskr stay A Section LG+F+R10 Location in the model tree ,Bootstraps It is displayed by the value on the branch .(C) Shared in Asgard virus VPF Membership ratio . In the legend of viral protein family classification A、B or E It means Archaea 、 Bacteria and / Or eukaryotic host .
We are Asgard UViGs Found in Caudovirales The marker protein of , Including small head protein 、 Base plate J、 Tail fibers 、 Portal protein and terminal enzyme large subunit ( chart 4). stay Ratatoskr、Nidhogg and Vedfolnir The putative capsid protein encoding the viral protein shell was found in . stay Ratatoskr I found a HK-97- Folded major capsid proteins (MCP). stay Nidhogg and Vedfolnir(PhANNs, Respectively 80% and 90% The degree of confidence ) Inferred detected in MCPs And contractile phage tail sheath like protein (>99.9% Probability ,e- value <4.4e-24) And phage procystin (99.5% Probability ,e- value 1.6e-13) There is structural similarity (HHPred, data S5).Fenrir and Ratatoskr All coding HNH Endonuclease , During the packaging process, you may DNA Split into units of genome length , And may work in coordination with their large subunit of terminating enzyme and portal protein . However , stay Fenrir and Sköll No putative MCPs, This may be due to the widespread presence of new viral structures in archaea specific viruses .MCPs The absence of may be due to capsid viruses and capsid free selfish elements (capsidless selfish elements,CSE) The close evolutionary connection . Archaeal viruses containing capsids acquire and lose capsid genes , Experienced to CSE Many changes back and forth , As seen in eukaryotic viruses , But it is still parasitic in the genetic information of its host .
To understand how Asgard viruses affect their hosts , We searched for prokaryotic viruses and NCLDVs Involves genome replication 、 Nucleotide metabolism 、 Transcription and host - Virus interacting genes ( chart 4). Asgard virus encodes the core that exists in prokaryotic and eukaryotic viruses DNA Copy genes (PolB、 ancient / Eukaryotic primer enzyme 、 Pincer loader 、RNase H and ATP Rely on the DNA Ligase )(26). It is predicted that ,Nidhogg Viruses have genes involved in autonomous genome replication and proofreading , Including a transcription factor S-II Zinc like finger domains ATP Dependence DNA Ligase (ligD)、RNase HI/ Reverse transcriptase like protein 、PolB And primer polymerase (PrimPol). It is predicted that , Viral PrimPol Like protein in DNA and / or RNA Primer activity has an additional role , And damage resistant DNA Polymerase activity . so far , These genes have not been described in archaeovirus , Only in Corynebacterium virus BFK20 and Acanthamoeba polyphaga ( Acanthamoeba )mimivirus The virus has been identified .Ratatoskr UViG Code an Archaea - Eukaryotes DNA Primer enzyme , be used for DNA Primer activity in replication , Also code PolB.Fenrir and Nidhogg code AAA ATP enzyme , And replication factors C Little Jackie (rfcS) And big subunit (rfcL) be similar , It may have the function of a pincer loader .Sköll There is proliferating cell nuclear antigen (PCNA), And from deep-sea sediments Lokiarchaeota Most closely related ( The protein similarity is 36.5%).PCNA It is an important replication process factor in eukaryotes and archaea , And exist in some of their viruses .Sköll It also encodes a ribonucleoside reductase ( Ribonucleoside diphosphate reductase ,β Subunits ) and hedgehog/intein Proteins with similar superfamily domains . Ribonucleoside reductase is encoded by a conserved core gene , be used for NCLDVs Nucleotide metabolism . There is one in this ribonucleotide reductase hedgehog/intein(Hint) Protein domains , It may indicate that this gene is transferred from the previous host to Sköll Of .Fenrir The virus may pass through a cell membrane during replication 5'(3') Halogen dehalogenase of deoxyribonuclease (HAD) Superfamily proteins to regulate dNTP pool .Fenrir It also contains a deoxynucleoside monophosphate kinase (DNMP kinase ), And deoxynucleoside monophosphate kinase of a large lake algae virus (bitscore=115.9) And a few Mimiviridae DNMP Kinases are similar .Fenrir、Nidhogg and Ratatoskr Viruses have DNA Repair genes , This is a NCLDVs One of the main features . This is in contrast to small viruses that lack these genes .Fenrir、Nidhogg and Ratatoskr The virus also encodes ERCC4 Endonuclease domain protein , With African swine fever endonuclease EP364R be similar .Nidhogg and Fenrir Coded ERCC4 Endonuclease and archaea XPF 3' Valve repair enzymes are similar . These similar XPF The endonuclease of contains helix - Hairpin - Helical domain , Be able to achieve non specificity DNA combination .
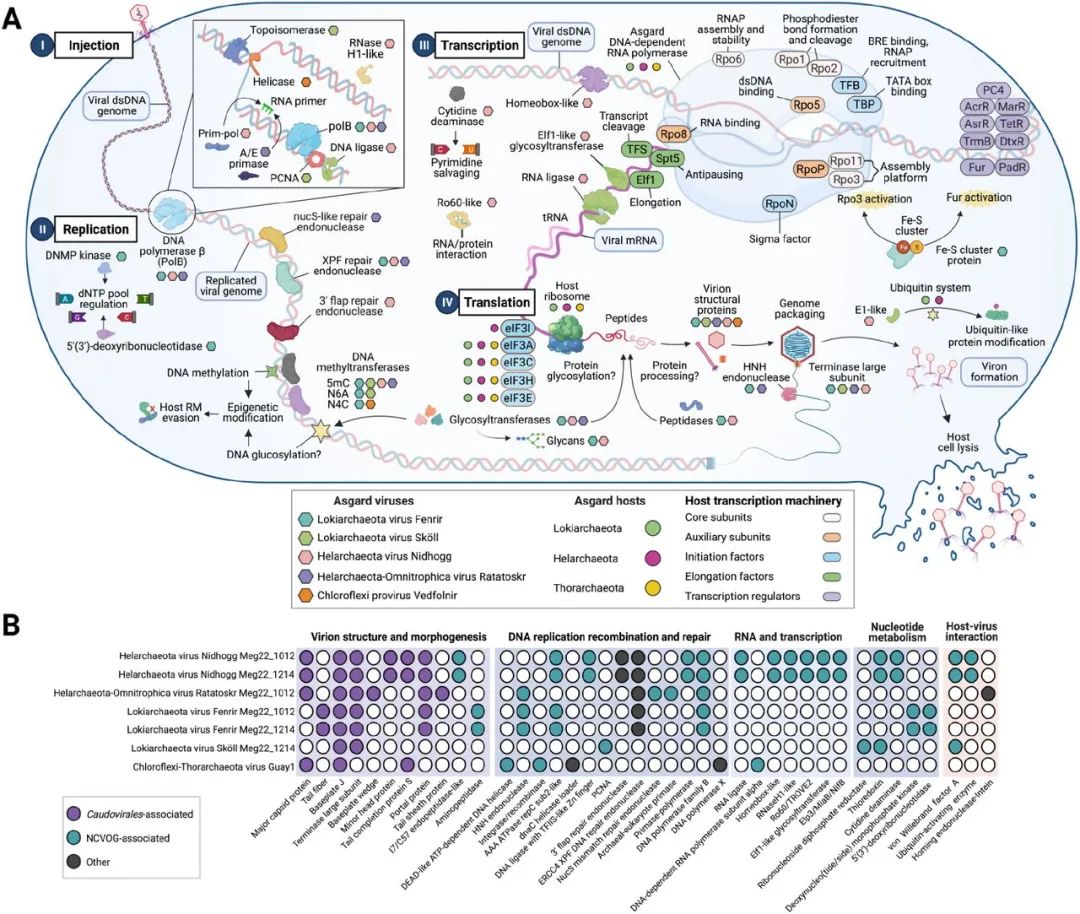
chart 4. The model of Asgard virus infection mechanism and its protein similarity with other viruses .
(A) Asgard virus is identified as involved in infection 、 Copy 、 Transcribed and translated genes . Asgard virus is shown as hexagonal in different colors , Asgard virus hosts are shown as circles of different colors , The host's transcription machine is marked by a functionally encoded ellipse ( See legend ).(B)40 A gene (X Lower part of shaft ) In Asgard virus (Y Axis ) Distribution in . Filled circles indicate annotated orthologues , The white circle indicates that no direct homologues have been found . abbreviation :NCVOG, Nucleoplasm giant DNA Viral protein homologues . use BioRender.com establish .
Except for the core DNA Copy machines and repair genes ,Fenrir and Nidhogg Viruses also have various mechanisms of gene expression and post-translational modification .Nidhogg Maybe through a homeodomain Proteins regulate transcription .Homeodomain The protein is DNA Binding transcription factors , It exists in eukaryotes and Mimiviridae in .Nidhogg The virus encodes a with TROVE and von Willebrand factor A Domain Ro60 protein .Ro60 May be related to non coding Y RNA Form ribonucleoprotein complex , With proteins and others RNA Interaction .Nidhogg The virus may also pass RNA Ligase repair 、 Splicing and editing transcripts ( chart 4).Fenrir Encode a winglike spiral - Turning helix protein , This is a common one found in archaeal virus transcription factors DNA combination motif. We are not in these UViG Any translation machine found in , This suggests that they utilize eukaryotic promoters present in host ribosomes and Asgard hosts . However ,Nidhogg and Fenrir It is possible to treat viral protein precursors with encoded peptidase . It is predicted that ,Nidhogg Encoded peptidase and Vaccinia Viruses (NCLDV A kind of virus )I7L/C57 The family of processed Endopeptidases is similar , and Fenrir It seems to produce M42 Family of aminopeptidases ( chart 4).
Several homologues of asgardman coding eukaryotic ubiquitin protein modification system . Eukaryotic viruses have been shown to hijack these systems to help in all stages of virus transmission .Nidhogg Encode three ubiquitin activation (ThiF family /E1-like) Protein homologues , You can use Helarchaeota Ubiquitin system (E1-like Activating enzyme 、E2-like Ligase and ubiquitin like protein domains ), And reuse it for replication 、 Transcription 、 The formation and release of virions . Yes Nidhogg Phylogenetic analysis of proteins showed , They are new E1 Ubiquitin activated homolog , yes Helarchaeota E1-like And eukaryotic ubiquitin like modifiers activate 5 The ancestor of protein . Given this type of host - Viral interactions have only been described in eukaryotic systems , This is very interesting. .
Asgard virus contains autonomous DNA Methylation and glycosylation genes , May play a role in host interactions .DNA Methylation has been recorded in bacteriophages and Mimiviridae in , As an epigenetic modification , Used to escape the host's defense , Such as restrictive modification (RM) System , Hijack the transcription machine , Adjust the replication cycle , and / Or defense has RM Competitive virus of the system . Interestingly ,Guaymas Asgard virus in the basin may be able to carry out autonomously DNA Methylation , Because they encode 5- Methylcytosine 、N4- Methylcytosine and D12 class N6- Methyladenine methyltransferase . Restriction endonuclease (RE) The simultaneous emergence of genes like , Such as Nidhogg Medium RecB family RE, May indicate functionality RM The system can inhibit the expression of host genes , And transfer the transcription from the host to the virus .Fenrir、Nidhogg and Ratatoskr It also encodes hexosyltransferase , This is a class of glycosyltransferases (GTs), It may be modified DNA、 The ability of hexose on protein or lipid , To escape the host defense system and inhibit the function of host molecules .GTs Already in bacteriophages and NCLDVs Confirmed in , But there is not enough record in archaeovirus .Fenrir Coding for two overlapping genes , Its sequence is similar to Sulfolobus islandicus rod-shaped virus 1 uncharacterized GT(SIRV1-GT) and alpha-1,2-mannosyltransferase(WbdA) Homology . This shows that Fenrir Can produce mannose sugar chain , These sugar chains are encoded SAM Dependent methyltransferase (WbdD) End , Instead of using the host machine . Besides ,Fenrir code L-malate GT(BshA), This is a Asgards Common genes in , It is believed to be involved in low molecular weight mercaptans in bacteria and archaea (LMWT) The biosynthesis of .Ratatoskr Code a N- acetyl -α-D- Glucosamine L- Malate dehydrogenase 1(BshB1) Homologues of , This gene is also associated with LMWT Related to the biosynthesis of . Last ,Nidhogg Code a with SIRV1-GT A match GT, And a glycogen synthase that may form glucan chains (GlgA) Homologues ( chart 4A).
In view of the relationship between Archaea asgardii and eukaryotes , Interestingly , Asgard infected virus has the same characteristics as eukaryotes NCLDV Similar replication and regulation mechanisms (PCNA、PolB、 Ribonucleotide reductase 、DNA Ligase 、DNMP Kinases and replication like factors C AAA ATP enzyme ). However , allow NCLDVs Decoupled transcription and Translation mRNA Capping genes do not seem to exist in these viruses . Besides , Many of their characteristics are different from those of archaeal viruses previously studied . for example , complete Nidhogg The genome encodes homeobox and PrimPol Protein domain , These protein domains are lacking in archaeal viruses . They also contain genes for ubiquitin protein modification systems , These genes have not been previously described in archaeoviruses , In eukaryotic viruses, it is used for virus reproduction . The structural proteins they encode are reminiscent of Caudovirales Those proteins in , However, most of their protein composition is different from that of this order . Asgard virus seems to have the viral characteristics of both Archaea and eukaryotes , This is consistent with the evolutionary position of their host . The first description of a virus associated with Archaea asgardii , It advances our understanding of the role of viruses in the ecology and evolution of Archaea asgardii .
reference
Rambo, I.M., Langwig, M.V., Leão, P. et al. Genomes of six viruses that infect Asgard archaea from deep-sea sediments. Nat Microbiol (2022). DOI: https://doi.org/10.1038/s41564-022-01150-8
Author's brief introduction

Brett J. Baker
Brett Baker He is an associate professor of marine microbiology in the Department of marine science and the Department of integrated biology at the University of Texas at Austin . His research aims to understand the ecology and evolution of uncultured bacteria and archaea . His laboratory mainly uses computational methods to describe the genomic diversity of marine sediments . In recent years , His team reconstructed the uncultured genome , For the first time, people have seen the physiological ability of new branches on the tree of life .Baker Doctor's research team also uses various activity-based methods to track the metabolic activities of these new microorganisms in nature . His laboratory has also played a role in characterizing Archaea related to eukaryotes , These Archaea have promoted our understanding of the origin of complex cell life .2014 year , He received a doctorate in Geological Sciences from the University of Michigan , There he studied the geological microbiology of deep-sea hydrothermal plumes and estuarine sediments . Before that , He worked at the University of California, Berkeley 10 Research assistant in , Study microorganisms related to acid mine drainage .
Guess you like
iMeta brief introduction Highly quoted articles High color value drawing imageGP Network analysis iNAP
iMeta Web tools Metabolism group MetOrigin Meggie cloud Lactation prediction DeepKla
iMeta review Intestinal flora Plant flora Oral flora Protein structure prediction
10000+: Flora analysis Babies and dogs Rhapsody of syphilis carry DNA Hair Nature
Series of tutorials : Introduction to microbiome Biostar Microbiome Macro genome
Expertise : Academic charts High score article Shengxin Scripture An indispensable person
Article to read : Macro genome Parasite benefits Evolutionary tree Necessary skills : put questions to Search for Endnote
Amplification analysis : Chart interpretation Analysis process Statistical mapping
Biological science : Intestinal bacteria Life on the human body Great leap forward in life Cell warfare The mystery of human body
Written in the back
To encourage readers to communicate and quickly solve scientific research difficulties , We established “ Macro genome ” Discussion groups , There are already domestic and foreign 6000+ Researchers join . Please add editor in chief wechat meta-genomics Bring you into the group , Be sure to remark “ full name - Company - Research direction - The title / grade ”. Please indicate your identity for the senior title , There are also microorganisms at home and abroad PI Cooperation and exchange of group supply . Ask for help with technical problems , First read 《 How to ask questions gracefully 》 Learn how to solve problems , Unresolved intra group discussions , Don't talk about problems in private , Help colleagues .
Click to read the original text , Jump to the latest article catalog to read
边栏推荐
- 【209】go语言的学习思想
- Halcon template matching
- I wrote a learning and practice tutorial for beginners!
- Li Kou brush question diary /day3/2022.6.25
- 【2022年江西省研究生数学建模】冰壶运动 思路分析及代码实现
- Crawler (6) - Web page data parsing (2) | the use of beautifulsoup4 in Crawlers
- 一、C语言入门基础
- 庆贺!科蓝SUNDB与中创软件完成七大产品的兼容性适配
- File processing examples of fopen, FREAD, fwrite, fseek
- 项目通用环境使用说明
猜你喜欢
I always thought that excel and PPT could only be used for making statements until I saw this set of templates (attached)
![[HCIA continuous update] network management and operation and maintenance](/img/a4/406b145793b701b001f04c7538dab3.png)
[HCIA continuous update] network management and operation and maintenance
12 - explore the underlying principles of IOS | runtime [isa details, class structure, method cache | t]

一种将Tree-LSTM的强化学习用于连接顺序选择的方法
TCP waves twice, have you seen it? What about four handshakes?
. Net ORM framework hisql practice - Chapter 2 - using hisql to realize menu management (add, delete, modify and check)
Make a grenade with 3DMAX
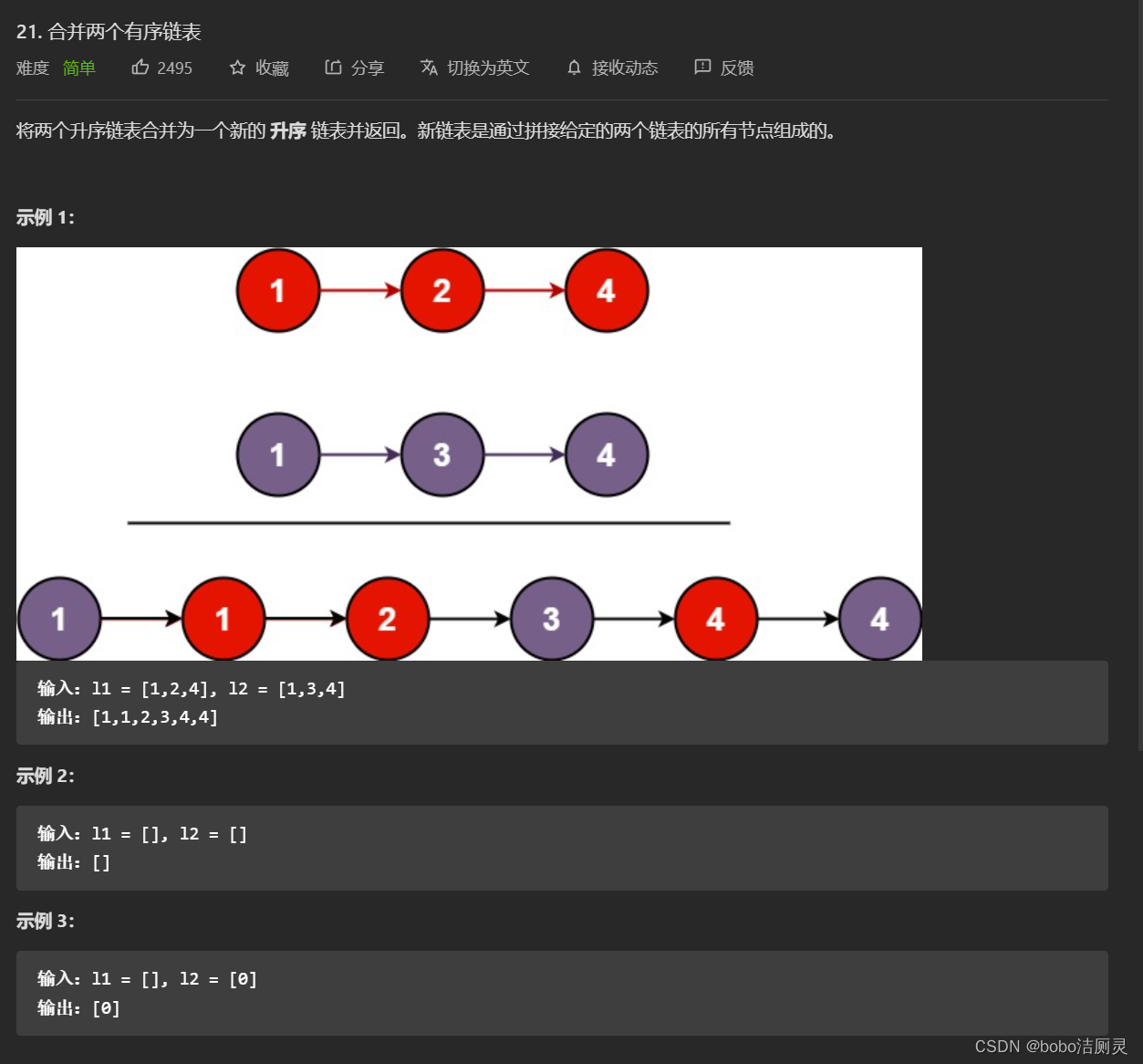
Li Kou brush question diary /day7/2022.6.29
Installation and use of VMware Tools and open VM tools: solve the problems of incomplete screen and unable to transfer files of virtual machines
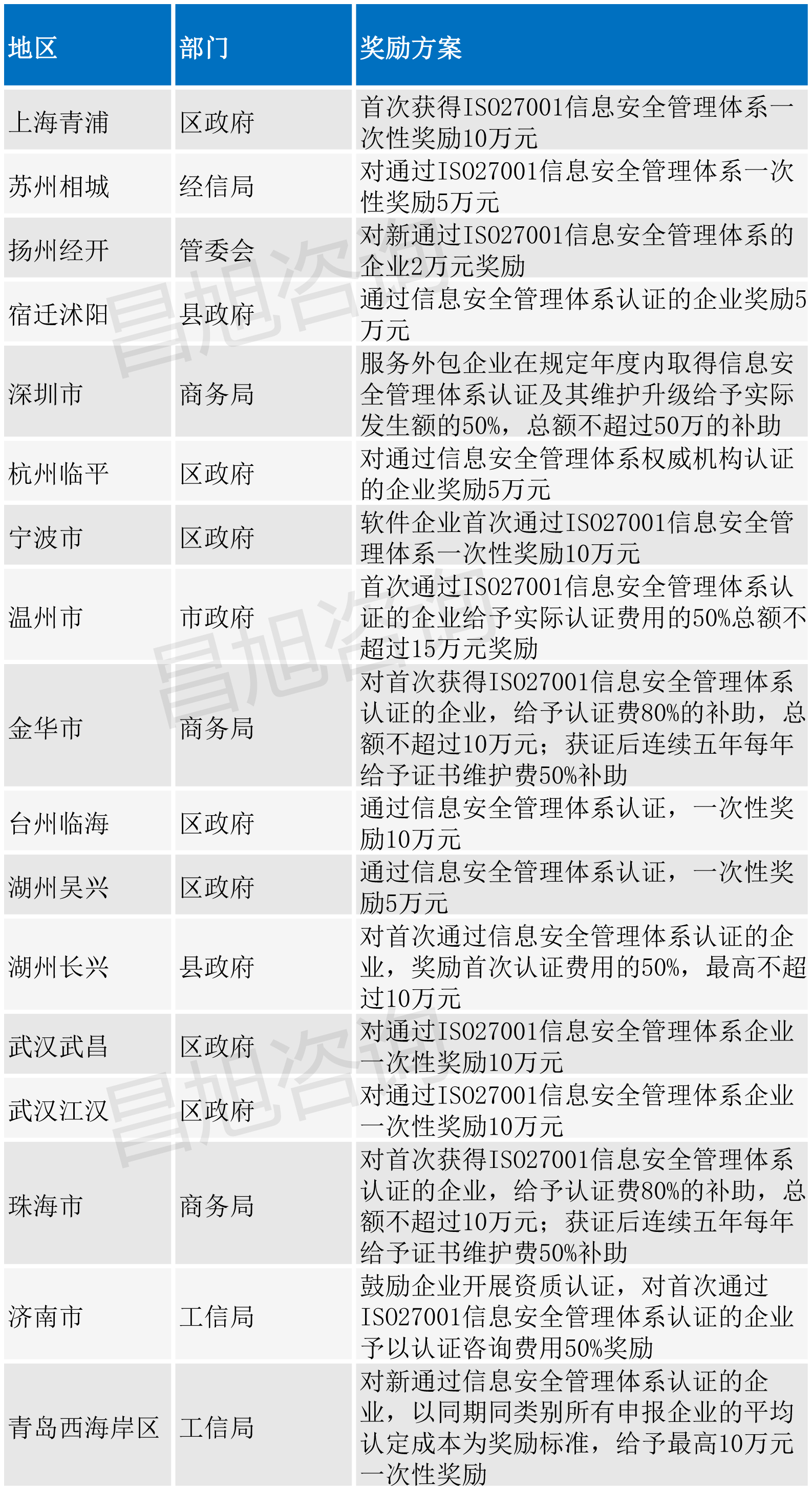
ISO27001认证办理流程及2022年补贴政策汇总
随机推荐
2022 national CMMI certification subsidy policy | Changxu consulting
中国农科院基因组所汪鸿儒课题组诚邀加入
The money circle boss, who is richer than Li Ka Shing, has just bought a building in Saudi Arabia
用于图数据库的开源 PostgreSQL 扩展 AGE被宣布为 Apache 软件基金会顶级项目
Li Kou brush question diary /day5/2022.6.27
删除二叉搜索树中的节点附图详解
Large scale service exception log retrieval
估值900亿,超级芯片IPO来了
ISO27001 certification process and 2022 subsidy policy summary
How to download files using WGet and curl
字节跳动Dev Better技术沙龙成功举办,携手华泰分享Web研发效能提升经验
【Go语言刷题篇】Go完结篇|函数、结构体、接口、错误入门学习
Li Kou brush question diary /day7/2022.6.29
[cloud native] what is the "grid" of service grid?
DB engines database ranking in July 2022: Microsoft SQL Server rose sharply, Oracle fell sharply
Lua EmmyLua 注解详解
股价大跌、市值缩水,奈雪推出虚拟股票,深陷擦边球争议
Mysql5.7 installation tutorial graphic explanation
Li Kou brush question diary /day7/6.30
[209] go language learning ideas
![12 - explore the underlying principles of IOS | runtime [isa details, class structure, method cache | t]](/img/c9/0feff5bb087bb082605cbf6cd1e194)