当前位置:网站首页>[Space & single cellomics] phase 1: single cell binding space transcriptome research PDAC tumor microenvironment
[Space & single cellomics] phase 1: single cell binding space transcriptome research PDAC tumor microenvironment
2022-07-02 14:37:00 【Top bioinformation】
**
6.19On that day, I released the creation on gongzong 「 Space & Literature study group of single cell omics 」 's post , At the same time, several thinking problems have been put forward . Because the number of people is controlled , The final team members only identified9 position, There are more than ten friends who contact me , I missed the bus , To you 「 sorry 」. If there are other groups in the future , I will inform you again . Study groupcourse、videoAnd so on and so on 「 Gongzong No. was released 」, You can pay attention to it .
Last Sunday , The group conducted The first 1 period Share , from TOP bacteria and Jeffery be the speaker , Respectively introduced :
2020 Single cell binding spatial transcriptome study PDAC Tumor microenvironment science Recently published single cell space metabonomics technology
This tweet yes TOP Bacteria literature sharing Text version , video Explanation already in B standing Release ; The... Of this issue 2 Tweets yes Jeffery Literature sharing Text version , Reprinted from his gongzong No : Student information programming self-study room
Let's go to the literature interpretation
background
 This article was published in 2020 year 1 month , It is the earliest single cell binding idling article I can find .
This article was published in 2020 year 1 month , It is the earliest single cell binding idling article I can find .
 The background of the article is mainly 3 spot :
The background of the article is mainly 3 spot :
scRNA-seq It will be dissociated before sequencing , Loss of spatial information Combining in situ hybridization and scRNA-seq It can solve the above problems to a certain extent , but ISH The limitations are also obvious , Only a small number of genes can be captured As early as 2016 In the year , Space transcriptome technology was born , But the limitation is the lack of single cell resolution
*This article combines scRNA-seq And spatial transcriptome , The two technologies complement each other , To explore the tumor microenvironment of pancreatic ductal adenocarcinoma
*
result
1. Identify cell subpopulations
 The technical route is relatively simple , The sample size of early articles is also small , The main content is based on the samples of two patients ( Paired samples are scarce )
The technical route is relatively simple , The sample size of early articles is also small , The main content is based on the samples of two patients ( Paired samples are scarce )
 First, we made cluster annotation for single cell data of two patients , Then I took a look at A,B Consistency of annotation results of two patient subgroups .
First, we made cluster annotation for single cell data of two patients , Then I took a look at A,B Consistency of annotation results of two patient subgroups .
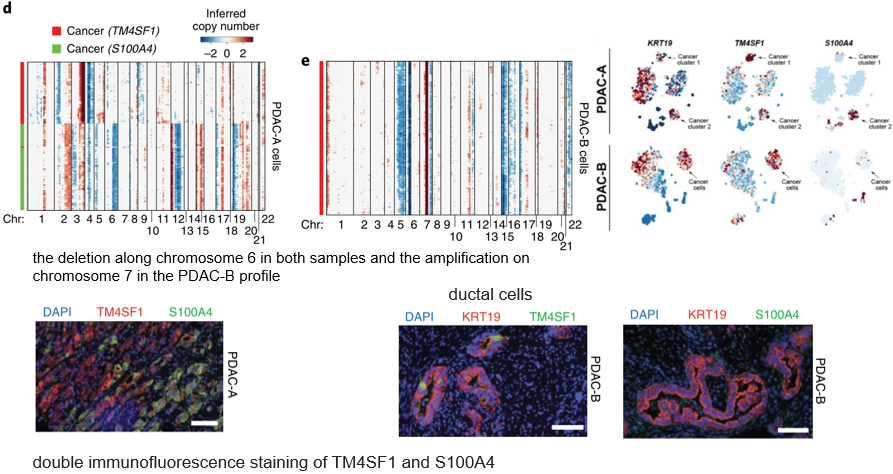 Still by inference CNV The malignant cells were identified by the method of , Further from CNV It can be seen from the heat map A There are two groups of patients CNV Cells with obvious differences . It was confirmed by immunofluorescence A,B The presence of tumor cells in the patient .
Still by inference CNV The malignant cells were identified by the method of , Further from CNV It can be seen from the heat map A There are two groups of patients CNV Cells with obvious differences . It was confirmed by immunofluorescence A,B The presence of tumor cells in the patient .
2. Analysis of spatial transcriptome data
 First of all, according to the HE Dyed pictures will A,B The patient's section is divided according to the tissue characteristics , From the spatial transcriptome data, we can also see some gene expression corresponding to the partition
First of all, according to the HE Dyed pictures will A,B The patient's section is divided according to the tissue characteristics , From the spatial transcriptome data, we can also see some gene expression corresponding to the partition
 Express data from idling only , Through the analysis process similar to single cell transcriptome , You can also see several groups spot, And it is consistent with the partition based on tissue slice
Express data from idling only , Through the analysis process similar to single cell transcriptome , You can also see several groups spot, And it is consistent with the partition based on tissue slice
3.MIA( Multimodal intersection analysis )
 In principle , Similar enrichment analysis , Is not complicated . The heat map on the right is viewed one by one , The deeper the red , It means that this cell type is in this region China and Vietnam are enriched . Through this analysis , Find out A Among patients , stay cancer region In addition to enrichment to tumor cells , It is also enriched in fibroblasts .
In principle , Similar enrichment analysis , Is not complicated . The heat map on the right is viewed one by one , The deeper the red , It means that this cell type is in this region China and Vietnam are enriched . Through this analysis , Find out A Among patients , stay cancer region In addition to enrichment to tumor cells , It is also enriched in fibroblasts .
4. Analysis of cell subclasses
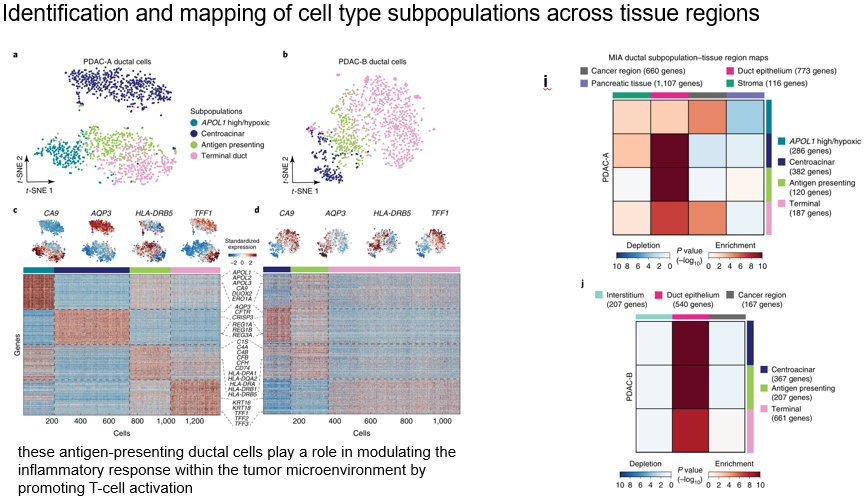
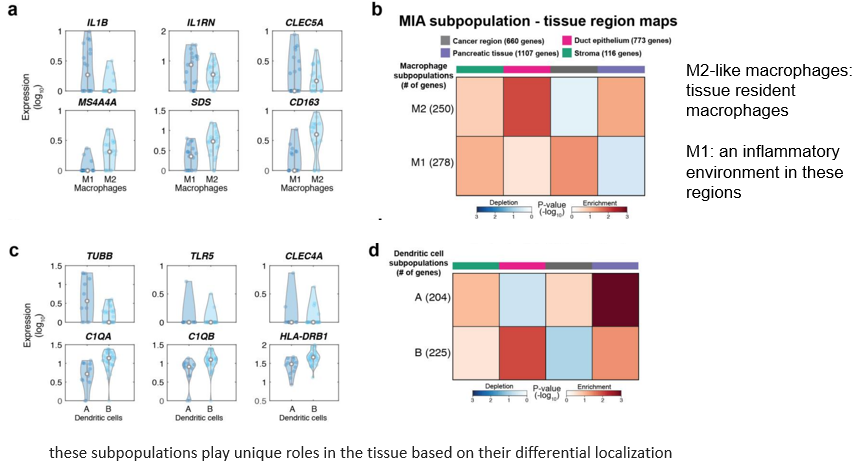 In this part, the ductal epithelial cells were analyzed in the same way 、 Macrophages 、DC cells . First, it is divided into sub categories , Reuse MIA Method to see the subclass in region The above enrichment .
In this part, the ductal epithelial cells were analyzed in the same way 、 Macrophages 、DC cells . First, it is divided into sub categories , Reuse MIA Method to see the subclass in region The above enrichment .
5. stay cancer region in , Whether different tumor cells co locate with different other cell types
 Take extra A Two sections of the patient , It is further subdivided in three slices cancer region. All three slices can be seen cancer cluster1 Co localization with fibroblasts .
Take extra A Two sections of the patient , It is further subdivided in three slices cancer region. All three slices can be seen cancer cluster1 Co localization with fibroblasts .
6. From the point of view of cell state scRNA-seq And idling
 utilize NMF From the single cell transcriptome data 3 Expression modules , The author focuses on stress-response Related modules .
utilize NMF From the single cell transcriptome data 3 Expression modules , The author focuses on stress-response Related modules . 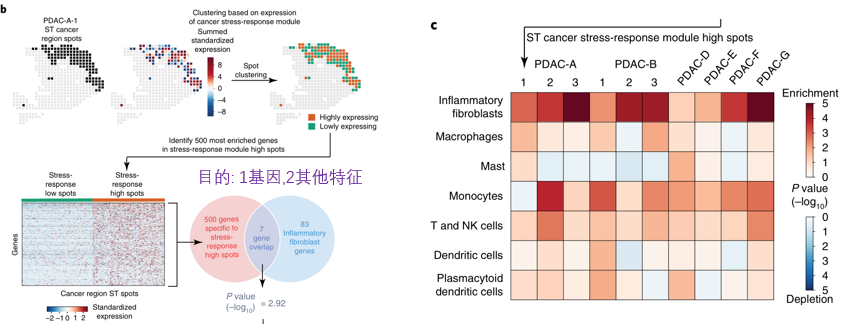 Then according to stress-response Module expression , take cancer region Of spot Divided into high and low groups , Continue the comparison between the two groups , find high This group of highly expressed genes . Obviously ,stress-response Related genes are already differential genes , Why do you do this ? There are at least two purposes for this :
Then according to stress-response Module expression , take cancer region Of spot Divided into high and low groups , Continue the comparison between the two groups , find high This group of highly expressed genes . Obviously ,stress-response Related genes are already differential genes , Why do you do this ? There are at least two purposes for this :
Increase the number of cells Reflect in addition stress-response Other features besides
The genes identified represent stress-response high Those of spot Regional characteristics of , And then use MIA Analysis stress-response high What cells will be enriched in the area of , Results show iCAF( Inflammatory fibroblasts ) Will be enriched in these areas .( There is a similar conclusion above , The conclusion here is to further refine .)
 TCGA Of bulk The sample also verified iCAF signature and stress-response module The relevance of
TCGA Of bulk The sample also verified iCAF signature and stress-response module The relevance of
This result was also confirmed by immunofluorescence .
The article also used mismatched melanoma samples , It goes further MIA Applicability of analysis .
summary
边栏推荐
- Delete element (with transition animation)
- < schematic diagram of oral arithmetic exercise machine program development> oral arithmetic exercise machine / oral arithmetic treasure / children's math treasure / children's calculator LCD LCD driv
- Fabric. JS zoom canvas
- MQ教程 | Exchange(交换机)
- Fabric.js 上划线、中划线(删除线)、下划线
- 篇9:XShell免费版安装
- Start to write a small demo - three piece chess
- 复用和分用
- Pychart connects to the remote server
- Go operation redis
猜你喜欢

Available solution development oral arithmetic training machine / math treasure / children's oral arithmetic treasure / intelligent math treasure LCD LCD driver ic-vk1622 (lqfp64 package), original te

HMS core machine learning service helps zaful users to shop conveniently
mathML转latex
Design and implementation of car query system based on php+mysql

途家木鸟美团夏日折扣对垒,门槛低就一定香吗?
![[to be continued] [UE4 notes] l5ue4 model import](/img/6b/d3083afc969043dbef1aeb4fccfc99.jpg)
[to be continued] [UE4 notes] l5ue4 model import
QT new project

Development and design of animation surrounding mall sales website based on php+mysql

Packet capturing tool Fiddler learning

Thoroughly master prototype__ proto__、 Relationship before constructor (JS prototype, prototype chain)
随机推荐
Fabric.js 上划线、中划线(删除线)、下划线
NLA自然语言分析,让数据分析更智能
Using computed in uni app solves the abnormal display of data () value in tab switching
Borui data integrated intelligent observable platform was selected into the "Yunyuan production catalogue" of China Academy of communications in 2022
Talk about idempotent design
Go operation redis
< schematic diagram of oral arithmetic exercise machine program development> oral arithmetic exercise machine / oral arithmetic treasure / children's math treasure / children's calculator LCD LCD driv
Check password
Threejs controller cube space basic controller + inertia control + flight control
taobao.trade.get( 获取单笔交易的部分信息),淘宝店铺订单接口,淘宝oAuth2.0接口,淘宝R2接口代码对接分享
The evolution process of the correct implementation principle of redis distributed lock and the summary of redison's actual combat
富文本编辑器添加矢量公式(MathType for TinyMCE ,可视化添加)
Packet capturing tool Fiddler learning
fatal: unsafe repository is owned by someone else 的解决方法
3. Function pointers and pointer functions
Essential elements of science fiction 3D scenes - City
Solving the longest subsequence with linear DP -- three questions
1. Editing weapon VIM
mongodb的认识
Some interview suggestions for Android programmers "suggestions collection"