当前位置:网站首页>How to Add P-Values onto Horizontal GGPLOTS
How to Add P-Values onto Horizontal GGPLOTS
2022-07-02 11:49:00 【Xiaoyu 2022】
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
# Box plots
bxp <- ggboxplot(df, x = "dose", y = "len", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
# Bar plots showing mean +/- SD
bp <- ggbarplot(df, x = "dose", y = "len", add = "mean_sd", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
stat.test <- df %>% t_test(len ~ dose)
# Box plot
stat.test <- stat.test %>% add_xy_position(x = "dose")
bxp + stat_pvalue_manual(stat.test, label = "p.adj.signif", tip.length = 0.01)
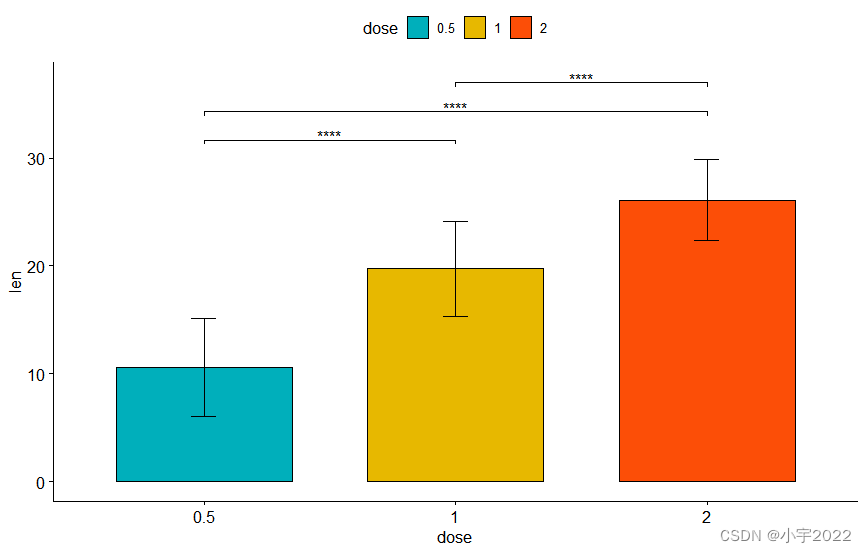
# Bar plot
stat.test <- stat.test %>% add_xy_position(fun = "mean_sd", x = "dose")
bp + stat_pvalue_manual(stat.test, label = "p.adj.signif", tip.length = 0.01)
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
# Box plots
bxp <- ggboxplot(df, x = "dose", y = "len", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
# Bar plots showing mean +/- SD
bp <- ggbarplot(df, x = "dose", y = "len", add = "mean_sd", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
stat.test <- stat.test %>% add_xy_position(x = "dose")
bxp +
stat_pvalue_manual(
stat.test, label = "p.adj.signif", tip.length = 0.01,
coord.flip = TRUE
) +
coord_flip()
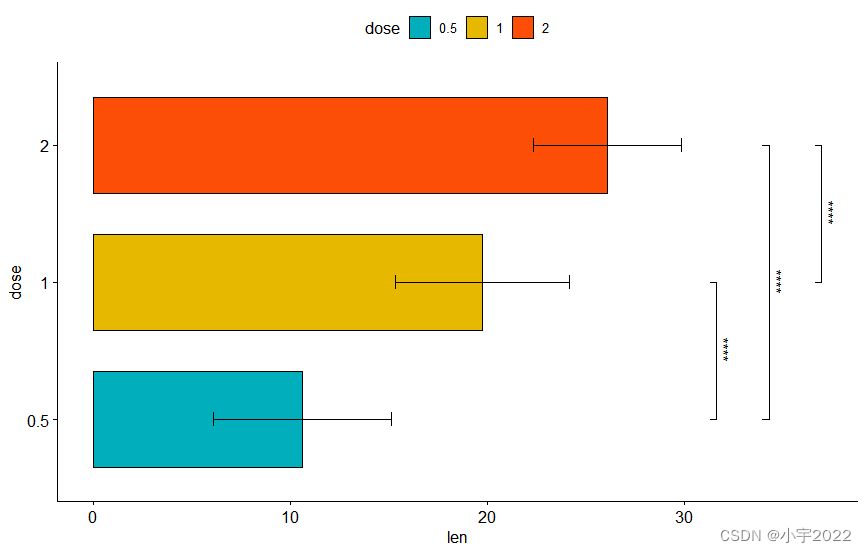
# Horizontal bar plot with p-values
stat.test <- stat.test %>% add_xy_position(fun = "mean_sd", x = "dose")
bp + stat_pvalue_manual(
stat.test, label = "p.adj.signif", tip.length = 0.01,
coord.flip = TRUE
) +
coord_flip()
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
# Box plots
bxp <- ggboxplot(df, x = "dose", y = "len", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
# Bar plots showing mean +/- SD
bp <- ggbarplot(df, x = "dose", y = "len", add = "mean_sd", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
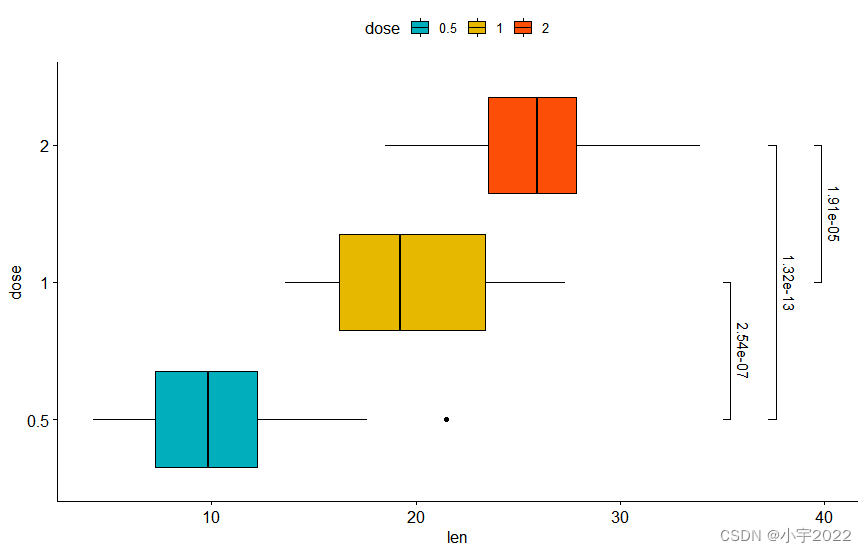
# Using `p.adj` as labels
stat.test <- stat.test %>% add_xy_position(x = "dose")
bxp +
stat_pvalue_manual(
stat.test, label = "p.adj", tip.length = 0.01,
coord.flip = TRUE
) +
coord_flip()
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
# Box plots
bxp <- ggboxplot(df, x = "dose", y = "len", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
# Bar plots showing mean +/- SD
bp <- ggbarplot(df, x = "dose", y = "len", add = "mean_sd", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
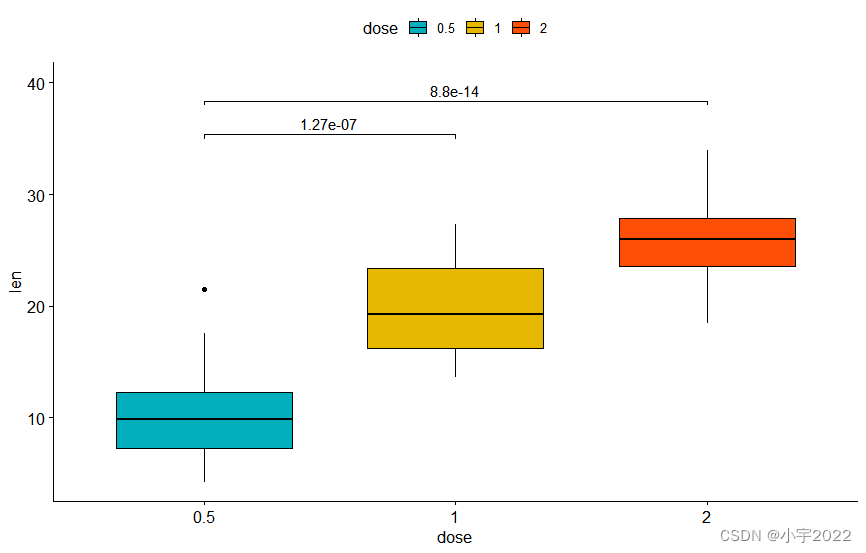
stat.test <- df %>% t_test(len ~ dose, ref.group = "0.5")
# Vertical box plot with p-values
stat.test <- stat.test %>% add_xy_position(x = "dose")
bxp +
stat_pvalue_manual(stat.test, label = "p.adj", tip.length = 0.01) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.1)))
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
# Box plots
bxp <- ggboxplot(df, x = "dose", y = "len", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
# Bar plots showing mean +/- SD
bp <- ggbarplot(df, x = "dose", y = "len", add = "mean_sd", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
stat.test <- df %>% t_test(len ~ dose, ref.group = "0.5")
# Vertical box plot with p-values
# Vertical bar plot with p-values
stat.test <- stat.test %>% add_xy_position(fun = "mean_sd", x = "dose")
bp +
stat_pvalue_manual(stat.test, label = "p.adj.signif", tip.length = 0.01) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.1)))
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
# Box plots
bxp <- ggboxplot(df, x = "dose", y = "len", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
# Bar plots showing mean +/- SD
bp <- ggbarplot(df, x = "dose", y = "len", add = "mean_sd", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
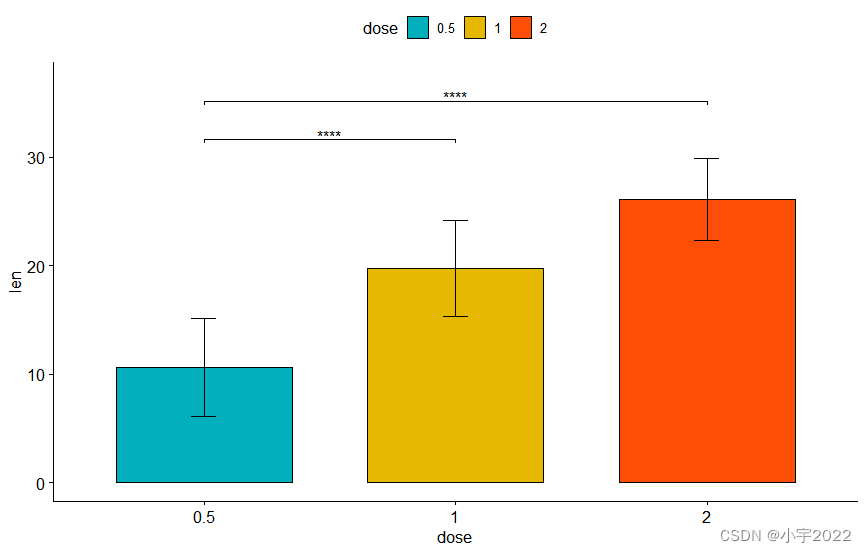
# Horizontal box plot with p-values
stat.test <- stat.test %>% add_xy_position(x = "dose")
bxp +
stat_pvalue_manual(
stat.test, label = "p.adj", tip.length = 0.01,
coord.flip = TRUE
) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.1))) +
coord_flip()

library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
# Box plots
bxp <- ggboxplot(df, x = "dose", y = "len", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
# Bar plots showing mean +/- SD
bp <- ggbarplot(df, x = "dose", y = "len", add = "mean_sd", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
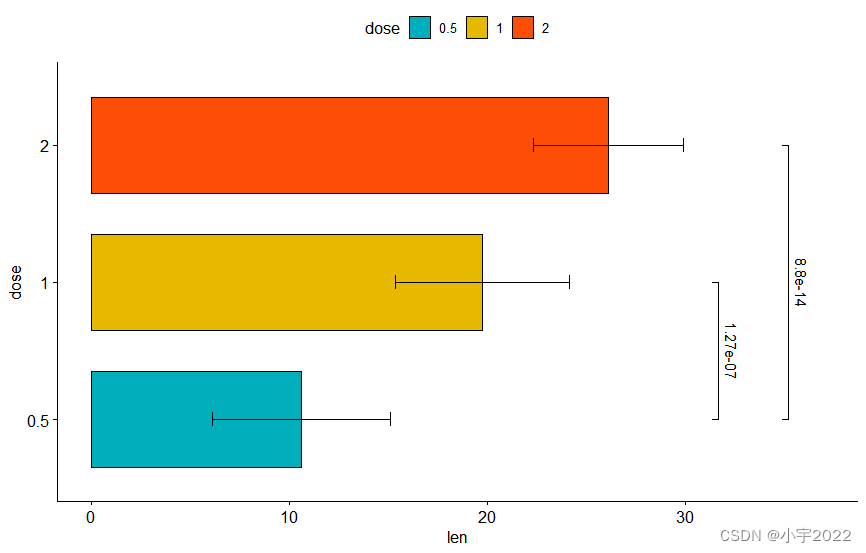
# Horizontal bar plot with p-values
stat.test <- stat.test %>% add_xy_position(fun = "mean_sd", x = "dose")
bp +
stat_pvalue_manual(
stat.test, label = "p.adj", tip.length = 0.01,
coord.flip = TRUE
) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.1))) +
coord_flip()
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
# Box plots
bxp <- ggboxplot(df, x = "dose", y = "len", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
# Bar plots showing mean +/- SD
bp <- ggbarplot(df, x = "dose", y = "len", add = "mean_sd", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
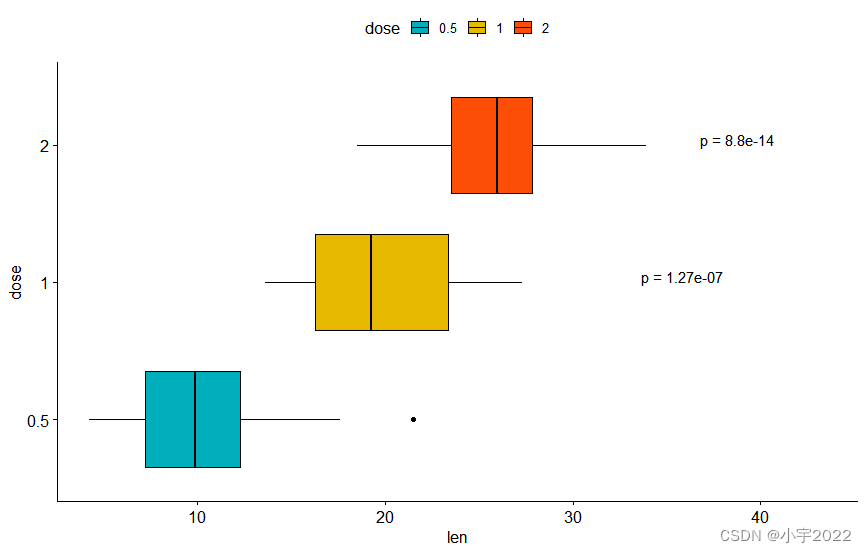
# Show p-values at x = "group2"
stat.test <- stat.test %>% add_xy_position(x = "dose")
bxp +
stat_pvalue_manual(
stat.test, label = "p = {p.adj}", tip.length = 0.01,
coord.flip = TRUE, x = "group2", hjust = 0.4
) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.2))) +
coord_flip()
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
# Box plots
bxp <- ggboxplot(df, x = "dose", y = "len", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
# Bar plots showing mean +/- SD
bp <- ggbarplot(df, x = "dose", y = "len", add = "mean_sd", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
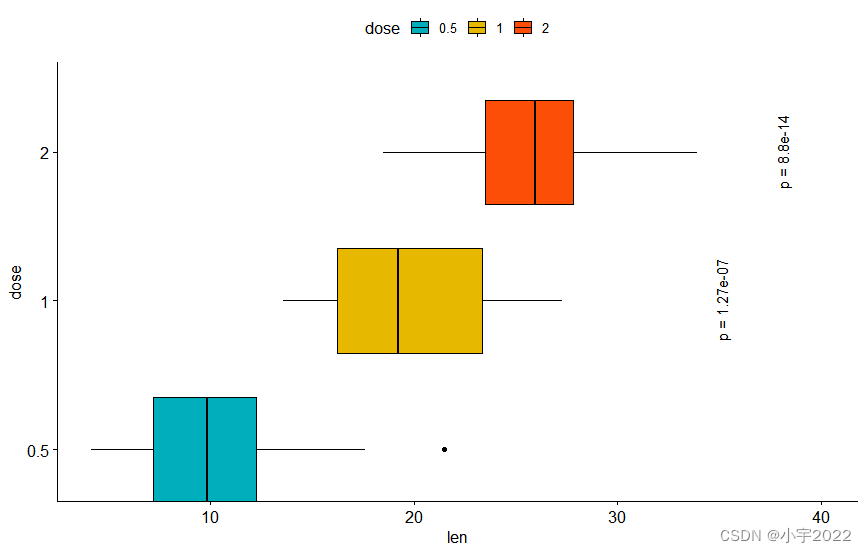
# Make the p-values horizontal using the option angle
stat.test <- stat.test %>% add_xy_position(x = "dose")
bxp +
stat_pvalue_manual(
stat.test, label = "p = {p.adj}", tip.length = 0.01,
coord.flip = TRUE, x = "group2", angle = 90
) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.1))) +
scale_x_discrete(expand = expansion(mult = c(0.05, 0.3))) +
coord_flip()
边栏推荐
- 电脑无缘无故黑屏,无法调节亮度。
- MySQL basic statement
- PYQT5+openCV项目实战:微循环仪图片、视频记录和人工对比软件(附源码)
- MySQL linked list data storage query sorting problem
- Power Spectral Density Estimates Using FFT---MATLAB
- Summary of data export methods in powerbi
- MySql存储过程游标遍历结果集
- vant tabs组件选中第一个下划线位置异常
- Beautiful and intelligent, Haval H6 supreme+ makes Yuanxiao travel safer
- 可升级合约的原理-DelegateCall
猜你喜欢
excel表格中选中单元格出现十字带阴影的选中效果
HOW TO CREATE AN INTERACTIVE CORRELATION MATRIX HEATMAP IN R

Always report errors when connecting to MySQL database

PowerBI中导出数据方法汇总
How to Visualize Missing Data in R using a Heatmap

JS -- take a number randomly from the array every call, and it cannot be the same as the last time

pgsql 字符串转数组关联其他表,匹配 拼接后原顺序展示
BEAUTIFUL GGPLOT VENN DIAGRAM WITH R
PYQT5+openCV项目实战:微循环仪图片、视频记录和人工对比软件(附源码)

6方面带你认识LED软膜屏 LED软膜屏尺寸|价格|安装|应用
随机推荐
TDSQL|就业难?腾讯云数据库微认证来帮你
Always report errors when connecting to MySQL database
CTF record
Pyqt5+opencv project practice: microcirculator pictures, video recording and manual comparison software (with source code)
PHP 2D and multidimensional arrays are out of order, PHP_ PHP scrambles a simple example of a two-dimensional array and a multi-dimensional array. The shuffle function in PHP can only scramble one-dim
Fabric.js 3个api设置画布宽高
bedtools使用教程
Principle of scalable contract delegatecall
由粒子加速器产生的反中子形成的白洞
RPA advanced (II) uipath application practice
Seriation in R: How to Optimally Order Objects in a Data Matrice
在连接mysql数据库的时候一直报错
ESP32音频框架 ESP-ADF 添加按键外设流程代码跟踪
Some suggestions for young people who are about to enter the workplace in the graduation season
GGHIGHLIGHT: EASY WAY TO HIGHLIGHT A GGPLOT IN R
The computer screen is black for no reason, and the brightness cannot be adjusted.
[multithreading] the main thread waits for the sub thread to finish executing, and records the way to execute and obtain the execution result (with annotated code and no pit)
RPA进阶(二)Uipath应用实践
CentOS8之mysql基本用法
Three transparent LED displays that were "crowded" in 2022