当前位置:网站首页>Learning DAVID Database (1)
Learning DAVID Database (1)
2022-07-31 03:38:00 【Huang Sibo】
Introduction:
DAIVD is a bioinformatics database and free online analysis software that helps users extract systematically comprehensive biological function annotation information (term) from large-scale gene or protein lists.Mainly used for differential gene function and pathway enrichment analysis.Web page: DAVID: Functional Annotation Result Summary
Official exercise data for users without gene numbers demolist1 and demolist2
https://david.ncifcrf.gov/helps/demo1.txthttps://david.ncifcrf.gov/helps/demo2.txtdemolist1 recorded up-regulation of 164 differentially expressed genes found in CD4+/CD62L-T cells relative to CD4+/CD62L-T cells

The content of the uploaded txt file can be gene ID or gene name. Different gene names are separated by newlines.
DAVID Gene ID Conversion Tool
Common genetic databases are Ensembl or NCBI
Enter gene ID: 3781 in the open NCBI web search box to find related gene information:
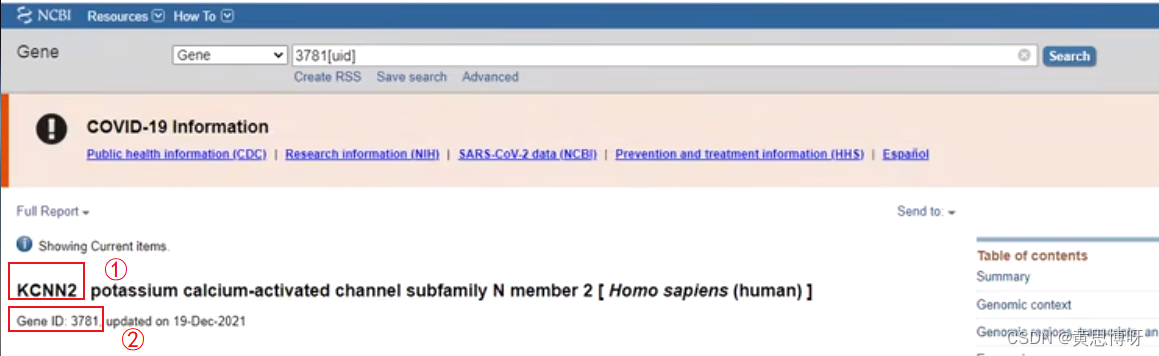
KCNN2 is the gene name and 3781 is the gene ID of KCNN2, both of which can be used as input data for DAVID
Upload bar action:
Copy the gene name or gene ID in the txt file to box A, or directly to the txt file
select identifier selection bar
entrez_gene_id: Gene ID corresponding to the number of uploaded dataofficial_gene_symbol : The corresponding uploaded data is the gene namestep3 Click Gene List
step4 Click Submit List , the page content is updated to step2:
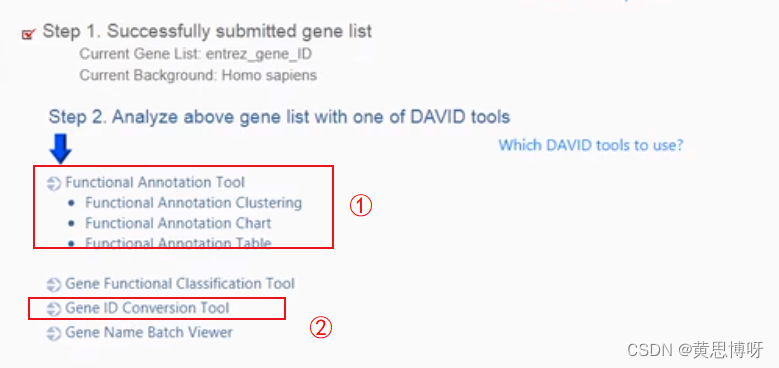
After submitting the gene list, step 2 uses multiple analysis directions, ①Functional Annotation Tool function for gene enrichment analysis, ②Gene ID Conversion Tool function for gene ID conversion, for example, convert the ID number of the NCBI gene library to the gene number of the Ensembl library (both correspond to the same gene)
Find species Latin name:
When performing gene ID conversion, you need to specify the species to be converted, and you need to enter the Latin name of the substance or the corresponding species ID number, such as human Latin homo sapiens, corresponding species ID: 9606
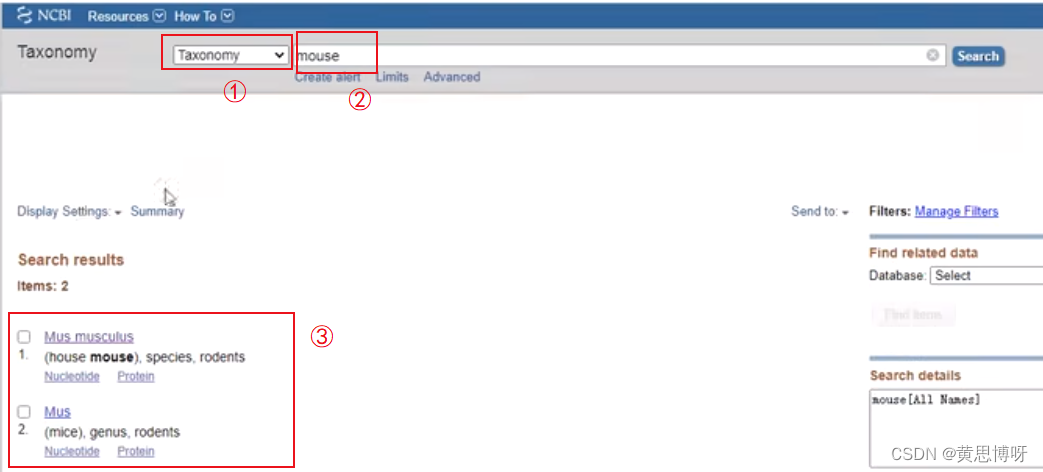
You can find the Latin name and corresponding species ID of the required species in the NCBI webpage
①Select the search range Taxomomy (taxonomy) from the drop-down box -->②Enter the English name of the target species, such as mouse mouse --> Click search -->③Show the matched species information
Gene ID conversion:

①There are 2500 gene IDs uploaded in the video, and 2355 human genes and 1 unknown gene are matched in the database
②Select the ID format you want to convert from the drop-down box, and select official_gene_symbol in the video, which is the official gene name
③Specify the species to which the gene belongs
④Click submit to convert and wait for the conversion to complete

①5 The status of conversion, successful indicates that the conversion is successful
②From column, is the uploaded genetic data
③To column, is the result of conversion, according to the selected official_gene_symbol, the converted gene name
④To column is just the abbreviation of gene name, David Gene Name column is the complete gene name
Click the Download File above to download the txt file and open it with Excel
Reference video link:
Introduction to gene enrichment tool DAVID (1)-Gene ID conversion_bilibili
边栏推荐
- errno error code and meaning (Chinese)
- 一份高质量的测试用例如何养成?
- LocalDate addition and subtraction operations and comparison size
- WebSocket Session is null
- 端口排查步骤-7680端口分析-Dosvc服务
- A brief introduction to the CheckboxListTile component of the basic components of Flutter
- BUG definition of SonarQube
- 浅识Flutter 基本组件之CheckboxListTile组件
- Safety 20220722
- Redis 使用 sorted set 做最新评论缓存
猜你喜欢
想从手工测试转岗自动化测试,需要学习哪些技能?

No qualifying bean of type question

Knowledge Distillation 7: Detailed Explanation of Knowledge Distillation Code

IIR filter and FIR filter
![[Dynamic programming] Maximum sum of consecutive subarrays](/img/3d/10731cc64d1c69d2beb3666ae0f064.png)
[Dynamic programming] Maximum sum of consecutive subarrays
![[Compilation principle] Lexical analysis program design principle and implementation](/img/eb/035234a402edf18beb7e2f82ebec17.png)
[Compilation principle] Lexical analysis program design principle and implementation

LocalDate addition and subtraction operations and comparison size
STM32 problem collection

$attrs/$listeners

web容器及IIS --- 中间件渗透方法1
随机推荐
MultipartFile file upload
TCP和UDP详解
Redis 使用LIST做最新评论缓存
想从手工测试转岗自动化测试,需要学习哪些技能?
Redis 使用 sorted set 做最新评论缓存
(五)final、抽象类、接口、内部类
Redis 统计用户新增和留存
3.5 】 【 Cocos Creator slow operating system to stop all animations
STM32 problem collection
分布式锁以及实现方式三种
some of my own thoughts
【Exception】The field file exceeds its maximum permitted size of 1048576 bytes.
Observer pattern
Distributed locks and three implementation methods
Mysql 45 study notes (23) How does MYSQL ensure that data is not lost
Based on the local, linking the world | Schneider Electric "Industrial SI Alliance" joins hands with partners to go to the future industry
(4) Recursion, variable parameters, access modifiers, understanding main method, code block
【CocosCreator 3.5】CocosCreator get network status
endian mode
What is a system?