当前位置:网站首页>/silicosis/geo/GSE184854_ scRNA-seq_ mouse_ lung_ ccr2/GSE184854_ RAW/GSM5598265_ matrix_ inflection_ demult
/silicosis/geo/GSE184854_ scRNA-seq_ mouse_ lung_ ccr2/GSE184854_ RAW/GSM5598265_ matrix_ inflection_ demult
2022-07-02 03:06:00 【youngleeyoung】
step1
library(Seurat)
#https://www.jianshu.com/p/5b26d7bc37b7
getwd()
path="G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/GSE184854_RAW/"
setwd(path)
getwd()
#new_counts=read.table(file = "G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/GSE184854_RAW/GSM5598265_matrix_inflection_demulti_SilicaWT.txt/GSM5598265_matrix_inflection_demulti_SilicaWT.txt")
head(new_counts)
# Or you can just create color urat object(counts=counts)
mydata <- CreateSeuratObject(counts = read.table(file = "G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/GSE184854_RAW/GSM5598265_matrix_inflection_demulti_SilicaWT.txt/GSM5598265_matrix_inflection_demulti_SilicaWT.txt"),
min.cells = 10, project = "mydata_scRNAseq")
dim(mydata)
Idents(mydata)
mydata$orig_stim=Idents(mydata)
table(Idents(mydata))
colnames(mydata)
library(stringr)
teststring=colnames(mydata)[1:50]
str_split(teststring,"_",simplify = T)
str_split(teststring,"_",simplify = T)[,2]
table(str_split(colnames(mydata),"_",simplify = T)[,2])
table(str_split(colnames(mydata),"_",simplify = T)[,1])
mydata$percent.mt=PercentageFeatureSet(mydata,pattern = "^Mt")
Idents(mydata)
grepl(colnames(mydata),pattern = 'A0301')
colnames(mydata)[grepl(colnames(mydata),pattern = 'A0301')]
table(Idents(mydata))
mydata=RenameIdents(mydata,'A0301'='day_21_1','A0302'='day_21_2','A0303'='day_21_3',
'A0304'='day_7_1','A0305'='day_7_2','A0306'='day_7_3',
'A0307'='day_3_1','A0308'='day_3_2','A0309'='day_3_3',
'doublet'='doublet')
mydata$mystim=Idents(mydata)
table(mydata$mystim)
dim(mydata)
Idents(mydata)=mydata$orig_stim
mydata=RenameIdents(mydata,'A0301'='Day_21','A0302'='Day_21','A0303'='Day_21',
'A0304'='Day_7','A0305'='Day_7','A0306'='Day_7',
'A0307'='Day_3','A0308'='Day_3','A0309'='Day_3',
'doublet'='Doublet')
mydata$my_stim_3=Idents(mydata)
save(mydata,file = "G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/GSE184854_RAW/mydata.rds")
step2
library(dplyr)
library(cowplot)
library(Seurat)
library(harmony)
getwd()
path="G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/GSE184854_RAW_all_merged/step_2_harmony/"
dir.create(path)
setwd(path)
getwd()
#load("G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/GSE184854_RAW/mydata.rds")
load("G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/GSE184854_RAW_all_merged/merged.rds")
table(Idents(All.merge))
All=subset(All.merge,idents = c('A0301', 'A0302', 'A0303',
'A0304', 'A0305' ,'A0306',
'A0307' , 'A0308', 'A0309'))
dim(All)
table(Idents(All))
#All$percent.mt=PercentageFeatureSet(All,pattern = "^mt-")
pdf("1_contorl_QC.pdf")
VlnPlot(All, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
dev.off()
All <- subset(All, subset = nFeature_RNA > 200 & percent.mt < 20)## 15499,16133;
All = All%>%Seurat::NormalizeData(verbose = FALSE) %>%
FindVariableFeatures(selection.method = "vst", nfeatures = 2000) %>%
ScaleData(verbose = FALSE)
All = RunPCA(All, npcs = 50, verbose = FALSE)
pdf("2_ElbowPlot.pdf")
ElbowPlot(All, ndims = 50)
dev.off()
table(Idents(All))
All$stim=Idents(All)
#All@meta.data$stim <- c(rep("case", length(grep("1$", All@assays$RNA@counts@Dimnames[[2]]))), rep("ctrl", length(grep("2$", All@assays$RNA@counts@Dimnames[[2]])))) ## 8186,7947;
pdf("2_pre_harmony_harmony_plot.pdf")
options(repr.plot.height = 5, repr.plot.width = 12)
p1 <- DimPlot(object = All, reduction = "pca", pt.size = .1, group.by = "stim")
p2 <- VlnPlot(object = All, features = "PC_1", group.by = "stim", pt.size = .1)
plot_grid(p1, p2)
dev.off()
##########################run harmony
All <- All %>% RunHarmony("stim", plot_convergence = TRUE)
harmony_embeddings <- Embeddings(All, 'harmony')
pdf("1_contorl_QC_.pdf")
VlnPlot(All, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
dev.off()
pdf("2_after_harmony_harmony_plot.pdf")
options(repr.plot.height = 5, repr.plot.width = 12)
p3 <- DimPlot(object = All, reduction = "harmony", pt.size = .1, group.by = "stim")
p4 <- VlnPlot(object = All, features = "harmony_1", group.by = "stim", pt.size = .1)
plot_grid(p3, p4)
dev.off()
#######################cluster
All <- All %>%
RunUMAP(reduction = "harmony", dims = 1:30) %>%
RunTSNE(reduction = "harmony", dims = 1:30) %>%
FindNeighbors(reduction = "harmony", dims = 1:30)
All<-All%>% FindClusters(resolution = 3) %>% identity()
options(repr.plot.height = 4, repr.plot.width = 10)
pdf("3_after_harmony_umap_two_group.pdf")
DimPlot(All, reduction = "umap", group.by = "stim", pt.size = .1)
dev.off()
pdf("3_after_harmony_cluster_UMAP.pdf")
DimPlot(All, reduction = "umap", label = TRUE, pt.size = .1)
dev.off()
pdf("3_umap_samples_split.pdf")
DimPlot(All, reduction = "umap", pt.size = .1, split.by = "stim", label = T)
dev.off()
pdf("3_after_harmony_tsne_two_group.pdf")
DimPlot(All, reduction = "tsne", group.by = "stim", pt.size = .1)
dev.off()
pdf("3_after_harmony_cluster_tSNE.pdf")
DimPlot(All, reduction = "tsne", label = TRUE, pt.size = .1)
dev.off()
pdf("3_tSNE_samples_split.pdf")
DimPlot(All, reduction = "tsne", pt.size = .1, split.by = "stim", label = T)
dev.off()
#######################################################################
stat = as.matrix(table(Idents(All), All$stim))
write.table(stat, "cluster_stat.txt", sep = "\t", quote = F, col.names = T, row.names = T)
################################################################
All<-All%>% FindClusters(resolution = 2) %>% identity()
Disease.markers <- FindAllMarkers(All, min.pct = 0.25, logfc.threshold = 0.35, only.pos = T)
head(Disease.markers)
top100markers <- Disease.markers %>% group_by(cluster) %>% slice_max(avg_log2FC,n=100)
openxlsx::write.xlsx(top100markers,'top100markers_for_all_res2.xlsx')
#write.table(top20markers, "top20_markers.txt", sep = "\t", quote = F, col.names = T, row.names = F)
#save(All, file = "G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/GSE184854_RAW_all_merged/step_2_harmony/all_harmony.rds")
load("G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/GSE184854_RAW_all_merged/step_2_harmony/all_harmony.rds")
getwd()
DimPlot(All,label = T)
FeaturePlot(All,features = c('Cd68'))
FeaturePlot(All,features = c('Lyz2'))
FeaturePlot(All,features = c('Mrc1'))
FeaturePlot(All,features = c('Adgre1'))
FeaturePlot(All,features = c('Axl'))
FeaturePlot(All,features = c('Csf1r'))
FeaturePlot(All,features = c('Ly6c2'))
FeaturePlot(All,features = c('Il7r'))
FeaturePlot(All,features = c('Car4'))
FeaturePlot(All,features = c('Siglecf'))
FeaturePlot(All,features = c('Mki67'))
FeaturePlot(All,features = c('Jchain','Tnfrsf17'),split.by = 'mystim')
DotPlot(All,features = c('Jchain','Tnfrsf17'),split.by = 'mystim')+RotatedAxis()+ggplot2::coord_flip()
colnames(All)
grepl(colnames(All),pattern = 'WT')
table(grepl(colnames(All),pattern = 'WT'))
table(grepl(colnames(All),pattern = 'CCR'))
All$orig_stim=Idents(All)
if(1==1){
All$mystim_validate=ifelse(grepl(colnames(All),pattern = 'WT') &
(grepl(colnames(All),pattern = 'A0301')|grepl(colnames(All),pattern = 'A0302')|grepl(colnames(All),pattern = 'A0303')),
paste0(colnames(All),'_Day_21_WT'),
ifelse(grepl(colnames(All),pattern = 'WT') &
(grepl(colnames(All),pattern = 'A0304')|grepl(colnames(All),pattern = 'A0305')|grepl(colnames(All),pattern = 'A0306'))
,paste0(colnames(All),'_Day_7_WT'),
ifelse(grepl(colnames(All),pattern = 'WT') &
(grepl(colnames(All),pattern = 'A0307')|grepl(colnames(All),pattern = 'A0308')|grepl(colnames(All),pattern = 'A0309'))
,paste0(colnames(All),'_Day_3_WT'),
ifelse(grepl(colnames(All),pattern = 'CCR') &
(grepl(colnames(All),pattern = 'A0301')|grepl(colnames(All),pattern = 'A0302')|grepl(colnames(All),pattern = 'A0303'))
,paste0(colnames(All),'_Day_21_CCR'),
ifelse(grepl(colnames(All),pattern = 'CCR') &
(grepl(colnames(All),pattern = 'A0304')|grepl(colnames(All),pattern = 'A0305')|grepl(colnames(All),pattern = 'A0306'))
,paste0(colnames(All),'_Day_7_CCR'),
ifelse(grepl(colnames(All),pattern = 'CCR') &
(grepl(colnames(All),pattern = 'A0307'))
,paste0(colnames(All),'_Day_3_CCR'),paste0(colnames(All),'_Day_3_CCR'))
)
))
)
)
All$mystim=ifelse(grepl(colnames(All),pattern = 'WT') &
(grepl(colnames(All),pattern = 'A0301')|grepl(colnames(All),pattern = 'A0302')|grepl(colnames(All),pattern = 'A0303')),
paste0('_Day_21_WT'),
ifelse(grepl(colnames(All),pattern = 'WT') &
(grepl(colnames(All),pattern = 'A0304')|grepl(colnames(All),pattern = 'A0305')|grepl(colnames(All),pattern = 'A0306'))
,paste0('_Day_7_WT'),
ifelse(grepl(colnames(All),pattern = 'WT') &
(grepl(colnames(All),pattern = 'A0307')|grepl(colnames(All),pattern = 'A0308')|grepl(colnames(All),pattern = 'A0309'))
,paste0('_Day_3_WT'),
ifelse(grepl(colnames(All),pattern = 'CCR') &
(grepl(colnames(All),pattern = 'A0301')|grepl(colnames(All),pattern = 'A0302')|grepl(colnames(All),pattern = 'A0303'))
,paste0('_Day_21_CCR'),
ifelse(grepl(colnames(All),pattern = 'CCR') &
(grepl(colnames(All),pattern = 'A0304')|grepl(colnames(All),pattern = 'A0305')|grepl(colnames(All),pattern = 'A0306'))
,paste0('_Day_7_CCR'),
ifelse(grepl(colnames(All),pattern = 'CCR') &
(grepl(colnames(All),pattern = 'A0307'))
,paste0('_Day_3_CCR'),paste0('_Day_3_CCR'))
)
))
)
)
}
table(All$mystim)
table(All$orig_stim)
table(All$orig.ident)
table(grep(colnames(All),pattern = 'WT') & (grep(colnames(All),pattern = 'A0301')|
grep(colnames(All),pattern = 'A0302')|
grep(colnames(All),pattern = 'A0303'))
)
All$mystim=ifelse(grepl(colnames(All),pattern = 'WT'),'WT','CCR')
table(All$mystim)
table(ifelse(grepl(colnames(All),pattern = 'WT'),'WT','CCR'))
length(grep(colnames(All),pattern = 'WT'))
table(All$orig.ident)
Idents(All)=All$mystim
table(grepl(colnames(All),pattern = 'WT') )
grepl(c('af','faf','cd'),pattern = 'a')
step3
getwd()
path="G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/step_3_mono_macrophage/"
dir.create(path)
setwd(path)
getwd()
load("G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/step_2_harmony/silicosis_mouse_harmony.rds")
table(Idents(All))
#csf1r and cd68
macrophage_and_monocyte=subset(All,idents =c('1','7','11','12','15','22',
'31','35') )
table(Idents(macrophage_and_monocyte))
DimPlot(macrophage_and_monocyte)
macrophage_and_monocyte$orig_idents_from_All=Idents(macrophage_and_monocyte)
table(Idents(macrophage_and_monocyte),macrophage_and_monocyte$stim)
subset_data=macrophage_and_monocyte
subset_data = subset_data %>%
Seurat::NormalizeData(verbose = FALSE) %>%
FindVariableFeatures(selection.method = "vst", nfeatures = 2000) %>%
ScaleData(verbose = FALSE) %>%
RunPCA(npcs = 50, verbose = FALSE)
ElbowPlot(subset_data, ndims = 50)
VlnPlot(subset_data, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
##########################run harmony
#BiocManager::install('harmony')
library('harmony')
subset_data <- subset_data %>% RunHarmony("stim", plot_convergence = TRUE)
harmony_embeddings <- Embeddings(subset_data, 'harmony')
#######################cluster
dims = 1:30
subset_data <- subset_data %>%
RunUMAP(reduction = "harmony", dims = dims) %>%
RunTSNE(reduction = "harmony", dims = dims) %>%
FindNeighbors(reduction = "harmony", dims = dims)
DimPlot(subset_data)
save(subset_data, file="sepsis_Endothelial.rds")
file=getwd()
r = 0.2
load(paste(file, "sepsis_Endothelial.rds", sep="/"))
dir.create(paste(file,r,sep="/"))
setwd(paste(file,r,sep="/"))
subset_data <- FindClusters(subset_data, resolution = r)
save(subset_data, file=paste0("sepsis_Endothelial_r", r, ".rds"))
subset_data <- ScaleData(subset_data, verbose = FALSE, features = rownames(subset_data))
pdf("1_Endothelial_cluster_TSNE.pdf")
p = DimPlot(subset_data, reduction = "tsne", label = TRUE, pt.size = 1,label.size = 6)
print(p)
dev.off()
pdf("1_Endothelial_cluster_UMAP.pdf")
p = DimPlot(subset_data, reduction = "umap", label = TRUE, pt.size = 1,label.size = 6)
print(p)
dev.off()
stat = cbind(table(Idents(subset_data), subset_data$stim), All = rowSums(table(Idents(subset_data), subset_data$stim)))
stat = rbind(stat, All = colSums(stat))
stat<-as.data.frame(stat)
stat
openxlsx::write.xlsx(stat, "2_Endothelial_cluster_stat.xlsx", col.names=T, row.names=T)
######################## find marker
markers <- FindAllMarkers(subset_data, min.pct = 0.25, logfc.threshold = 0.25, only.pos=T)
openxlsx::write.xlsx(markers,"3_Endothelial_cluster_markers.xlsx", col.names=T,row.names=F)
FeaturePlot(subset_data,features = c('C1qa',
'C1qb',
'C1qc'))
FeaturePlot(subset_data,features = c('Mmp9',
'Spp1',
'Marcks',
'Il1rn'))
FeaturePlot(subset_data,features = c('Mmp9',
'Spp1',
'Marcks',
'Il1rn'),split.by = 'stim')
DotPlot(subset_data,features = c('Mmp9',
'Spp1',
'Marcks',
'Il1rn',
'C1qa','C1qb','C1qc'))
r=0.3
load(paste(file, "sepsis_Endothelial.rds", sep="/"))
dir.create(paste(file,r,sep="/"))
setwd(paste(file,r,sep="/"))
subset_data <- FindClusters(subset_data, resolution = r)
save(subset_data, file=paste0("sepsis_Endothelial_r", r, ".rds"))
subset_data <- ScaleData(subset_data, verbose = FALSE, features = rownames(subset_data))
pdf("1_Endothelial_cluster_TSNE.pdf")
p = DimPlot(subset_data, reduction = "tsne", label = TRUE, pt.size = 1,label.size = 6)
print(p)
dev.off()
pdf("1_Endothelial_cluster_UMAP.pdf")
p = DimPlot(subset_data, reduction = "umap", label = TRUE, pt.size = 1,label.size = 6)
print(p)
dev.off()
stat = cbind(table(Idents(subset_data), subset_data$stim), All = rowSums(table(Idents(subset_data), subset_data$stim)))
stat = rbind(stat, All = colSums(stat))
stat<-as.data.frame(stat)
stat
openxlsx::write.xlsx(stat, "2_Endothelial_cluster_stat.xlsx", col.names=T, row.names=T)
######################## find marker
markers <- FindAllMarkers(subset_data, min.pct = 0.25, logfc.threshold = 0.25, only.pos=T)
openxlsx::write.xlsx(markers,"3_Endothelial_cluster_markers.xlsx", col.names=T,row.names=F)
DimPlot(subset_data,label = T)
step4
getwd()
path="G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/step_4_macrophage_from_0.3"
dir.create(path)
setwd(path)
getwd()
load("G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/step_3_mono_macrophage/0.3/sepsis_Endothelial_r0.3.rds")
DimPlot(subset_data,label = T)
subset_data=subset(subset_data,idents = c('1','2','4'))
DimPlot(subset_data,label = T)
subset_data$cluster124=Idents(subset_data)
# Merge
subset_data = subset_data %>%
Seurat::NormalizeData(verbose = FALSE) %>%
FindVariableFeatures(selection.method = "vst", nfeatures = 2000) %>%
ScaleData(verbose = FALSE) %>%
RunPCA(npcs = 50, verbose = FALSE)
ElbowPlot(subset_data, ndims = 50)
VlnPlot(subset_data, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
##########################run harmony
#BiocManager::install('harmony')
library('harmony')
subset_data <- subset_data %>% RunHarmony("stim", plot_convergence = TRUE)
harmony_embeddings <- Embeddings(subset_data, 'harmony')
#######################cluster
dims = 1:30
subset_data <- subset_data %>%
RunUMAP(reduction = "harmony", dims = dims) %>%
RunTSNE(reduction = "harmony", dims = dims) %>%
FindNeighbors(reduction = "harmony", dims = dims)
DimPlot(subset_data)
FeaturePlot(subset_data,features = c('C1qa',
'C1qb',
'C1qc'))
FeaturePlot(subset_data,features = c('Mmp9',
'Spp1',
'Marcks',
'Il1rn'))
FeaturePlot(subset_data,features = c('Mmp9',
'Spp1',
'Marcks',
'Il1rn'),split.by = 'stim')
DotPlot(subset_data,features = c('Mmp9',
'Spp1',
'Marcks',
'Il1rn',
'C1qa','C1qb','C1qc',
'Csf1','Csf1r',
'Car4', 'Ctsk', 'Chil3', 'S100a1','Wfdc21',
'Ear1', 'Fabp1',
'Itgam', 'Cd36','Gpnmb',
'Litaf', 'Jund', 'Bhlhe40', 'Bhlhe41','Klf9'))
subset_data=RenameIdents(subset_data,'4'='IM','2'='AM','1'='Mo-AM')
getwd()
#save(subset_data,file = "G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/step_4_macrophage_from_0.3/im_am_from_res0.3.rds")
load("G:/silicosis/geo/GSE184854_scRNA-seq_mouse_lung_ccr2/step_4_macrophage_from_0.3/im_am_from_res0.3.rds")
library(Seurat)
DotPlot(subset_data,features = c('Chi3l1', 'Marcks', 'Il1rn', 'Pla2g7', 'Mmp9', 'Spp1',
'Mmp12','Mmp14'))
DotPlot(All.merge,features = c('Spp1','Mmp14','Il1rn','Marcks','Pla2g7'))
VlnPlot(subset_data,features = c('Chi3l1', 'Marcks', 'Il1rn', 'Pla2g7', 'Mmp9', 'Spp1',
'Mmp12','Mmp14'))
#'Apoe', 'Mafb',
DotPlot(subset_data,features = c('C1qa','C1qb','C1qc',
'Ear1','Fabp1', 'Ctsk','Car4', 'Chil3', 'S100a1' , 'Wfdc21',
'Itgam', 'Litaf', 'Gpnmb','Apoe','Mafb',
'Mmp14','Spp1'))+RotatedAxis()+ggplot2:::coord_flip()
DotPlot(All.merge,features = c('Il34','Csf1'))
VlnPlot(All.merge,features = 'Il34')
VlnPlot(All.merge,features = 'Csf1')
VlnPlot(All.merge,features = c('Pdgfa','App','Tgfb1'))
DotPlot(All.merge,features = c('Il34','Csf1','Fgf2'))
边栏推荐
- Set status bar color
- QT environment generates dump to solve abnormal crash
- verilog 并行块实现
- Competition and adventure burr
- 3048. Number of words
- PHP notes - use Smarty to set public pages (include, if, else, variable settings)
- [staff] the direction of the symbol stem and the connecting line (the symbol stem faces | the symbol stem below the third line faces upward | the symbol stem above the third line faces downward | the
- SAP ui5 beginner tutorial 19 - SAP ui5 data types and complex data binding
- 多线程查询,效率翻倍
- Golang configure export goprivate to pull private library code
猜你喜欢

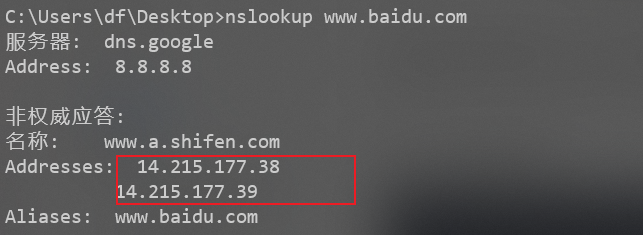
About DNS
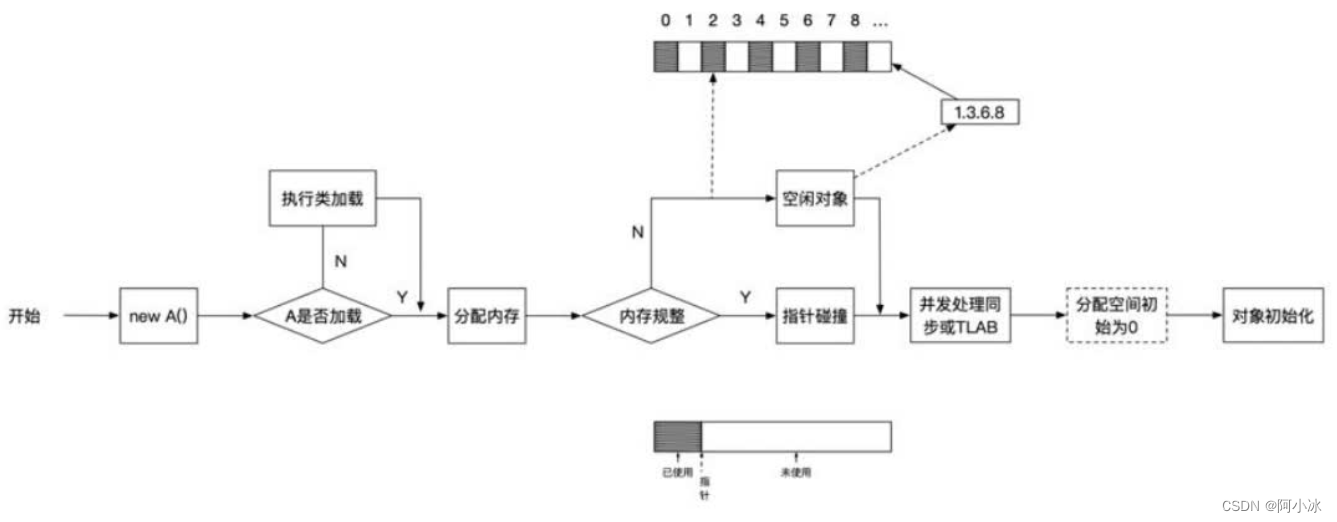
【JVM】创建对象的流程详解

結婚後

Share the basic knowledge of a common Hongmeng application

STM32__ 05 - PWM controlled DC motor

2022-2028 global encryption software industry research and trend analysis report

Is bone conduction earphone better than traditional earphones? The sound production principle of bone conduction earphones is popular science

AcWing 245. Can you answer these questions (line segment tree)

MongoDB非关系型数据库
批量检测url是否存在cdn—高准确率
随机推荐
[JS reverse series] analysis of a customs publicity platform
竞争与冒险 毛刺
What is the principle of bone conduction earphones and who is suitable for bone conduction earphones
2022-2028 global human internal visualization system industry research and trend analysis report
结婚后
命名块 verilog
Mmsegmentation series training and reasoning their own data set (3)
Calculation of page table size of level 2, level 3 and level 4 in protection mode (4k=4*2^10)
ZABBIX API creates hosts in batches according to the host information in Excel files
Mathematical calculation in real mode addressing
IPhone 6 plus is listed in Apple's "retro products" list
OSPF LSA message parsing (under update)
32, 64, 128 bit system
Basic 01: print string
Just a few simple steps - start playing wechat applet
Jvm-01 (phased learning)
超图iServer rest服务之feature查询
The number one malware in January 2022: lokibot returned to the list, and emotet returned to the top
Cache processing scheme in high concurrency scenario
创业了...