当前位置:网站首页>HOW TO ADD P-VALUES TO GGPLOT FACETS
HOW TO ADD P-VALUES TO GGPLOT FACETS
2022-07-02 11:49:00 【Xiaoyu 2022】
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
stat.test <- df %>%
group_by(dose) %>%
t_test(len ~ supp) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance()
# Create a box plot
bxp <- ggboxplot(
df, x = "supp", y = "len", fill = "#00AFBB",
facet.by = "dose"
)
# Make facet and add p-values
stat.test <- stat.test %>% add_xy_position(x = "supp")
bxp + stat_pvalue_manual(stat.test)

library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
stat.test <- df %>%
group_by(dose) %>%
t_test(len ~ supp) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance()
# Create a box plot
bxp <- ggboxplot(
df, x = "supp", y = "len", fill = "#00AFBB",
facet.by = "dose"
)
# Make facet and add p-values
stat.test <- stat.test %>% add_xy_position(x = "supp")
bxp + stat_pvalue_manual(stat.test)
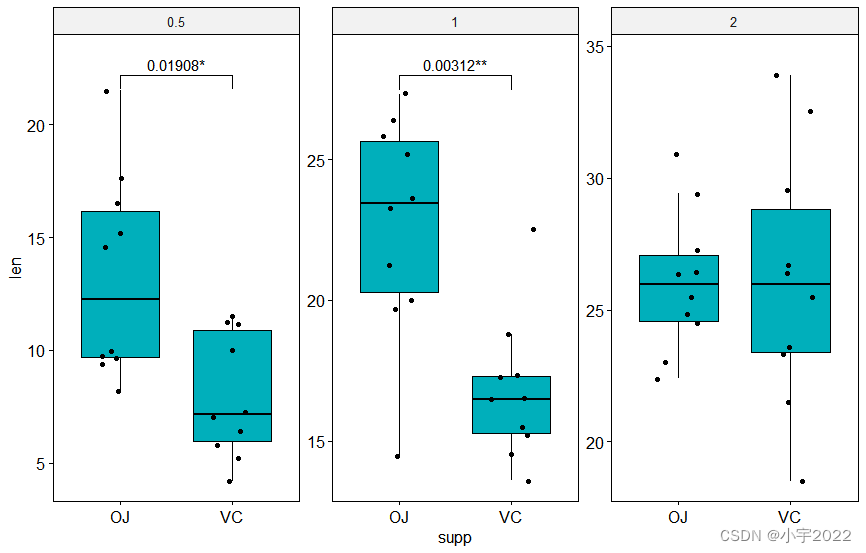
# Make the facet scale free and add jitter points
# Move down the bracket using `bracket.nudge.y`
# Hide ns (non-significant)
# Show adjusted p-values and significance levels
# Add 10% spaces between the p-value labels and the plot border
bxp <- ggboxplot(
df, x = "supp", y = "len", fill = "#00AFBB",
facet.by = "dose", scales = "free", add = "jitter"
)
bxp +
stat_pvalue_manual(
stat.test, bracket.nudge.y = -2, hide.ns = TRUE,
label = "{p.adj}{p.adj.signif}"
) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.1)))
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
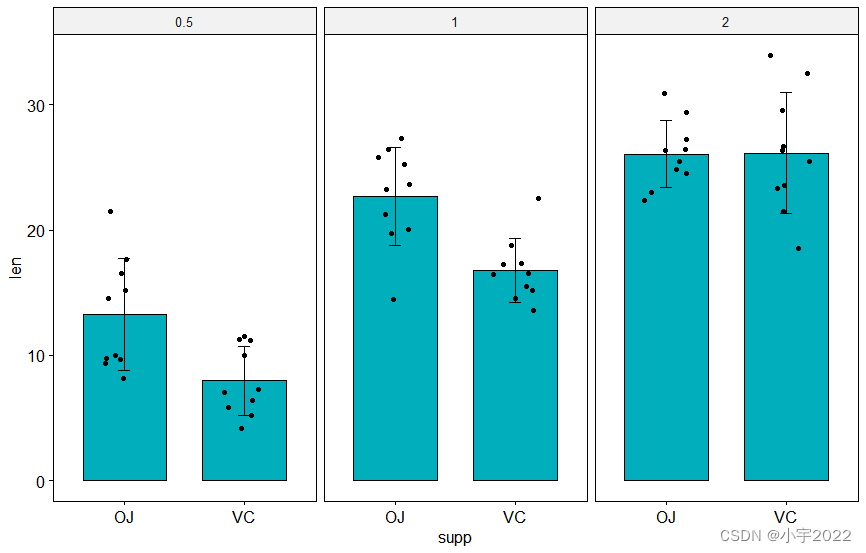
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = "mean_sd",
fill = "#00AFBB", facet.by = "dose"
)
# Add p-values onto the bar plots
stat.test <- stat.test %>% add_xy_position(fun = "mean_sd", x = "supp")
bp + stat_pvalue_manual(stat.test)

library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
# Add p-values onto the bar plots
stat.test <- stat.test %>% add_xy_position(fun = "max", x = "supp")
bp + stat_pvalue_manual(stat.test)

library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
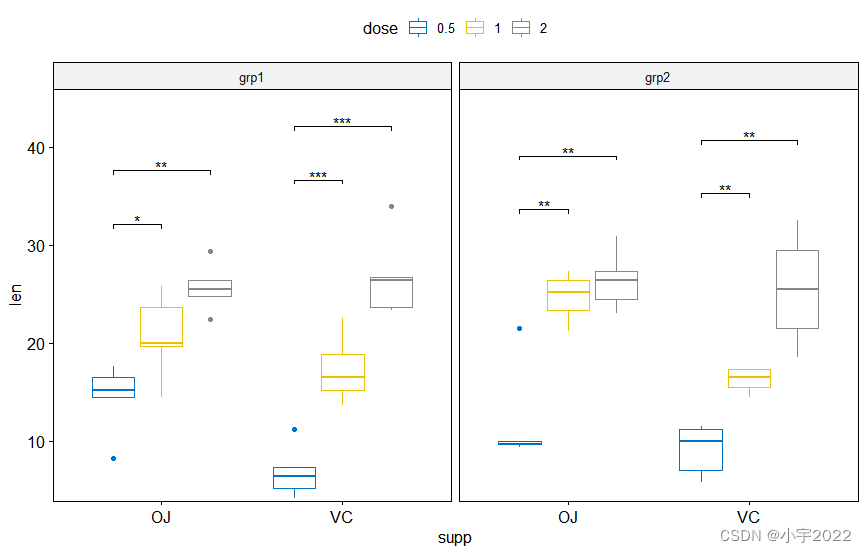
stat.test <- df %>%
group_by(group, dose) %>%
t_test(len ~ supp) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance()
# Create box plots with significance levels
# Hide ns (non-significant)
stat.test <- stat.test %>% add_xy_position(x = "supp")
ggboxplot(
df, x = "supp", y = "len", fill = "#E7B800",
facet = c("group", "dose")
) +
stat_pvalue_manual(stat.test, hide.ns = TRUE)

library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
stat.test <- df %>%
group_by(group, dose) %>%
t_test(len ~ supp) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance()
# Create bar plots with significance levels
# Hide ns (non-significant)
stat.test <- stat.test %>% add_xy_position(x = "supp", fun = "mean_sd")
ggbarplot(
df, x = "supp", y = "len", fill = "#E7B800",
add = c("mean_sd", "jitter"), facet = c("group", "dose")
) +
stat_pvalue_manual(stat.test, hide.ns = TRUE)

library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
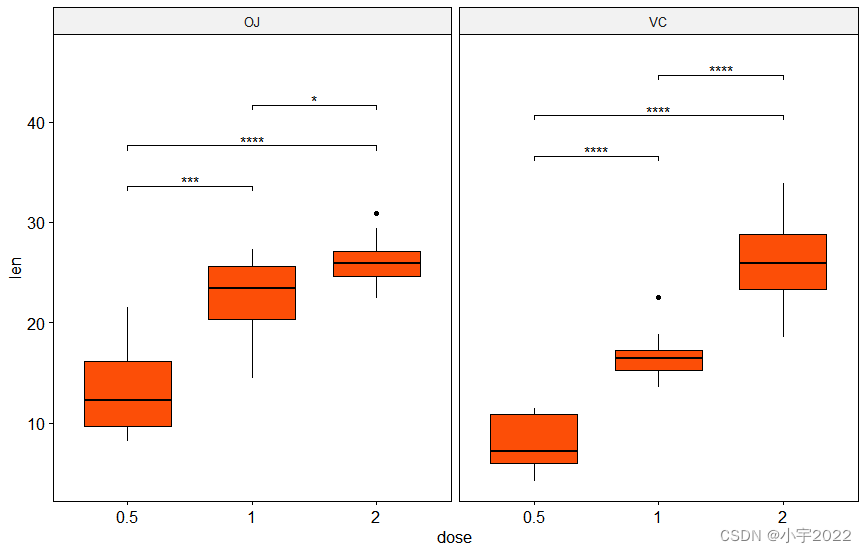
stat.test <- df %>%
group_by(supp) %>%
t_test(len ~ dose)
# Box plots with p-values
stat.test <- stat.test %>% add_y_position()
ggboxplot(df, x = "dose", y = "len", fill = "#FC4E07", facet.by = "supp") +
stat_pvalue_manual(stat.test, label = "p.adj.signif", tip.length = 0.01) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.1)))
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
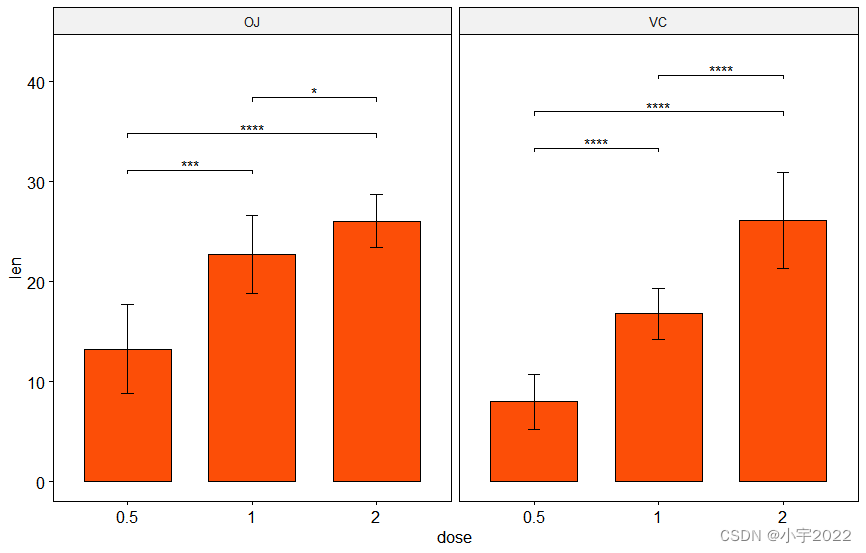
# Bar plot with p-values
# Add 10% space on the y-axis above the box plots
stat.test <- stat.test %>% add_y_position(fun = "mean_sd")
ggbarplot(
df, x = "dose", y = "len", fill = "#FC4E07",
add = "mean_sd", facet.by = "supp"
) +
stat_pvalue_manual(stat.test, label = "p.adj.signif", tip.length = 0.01) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.1)))
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
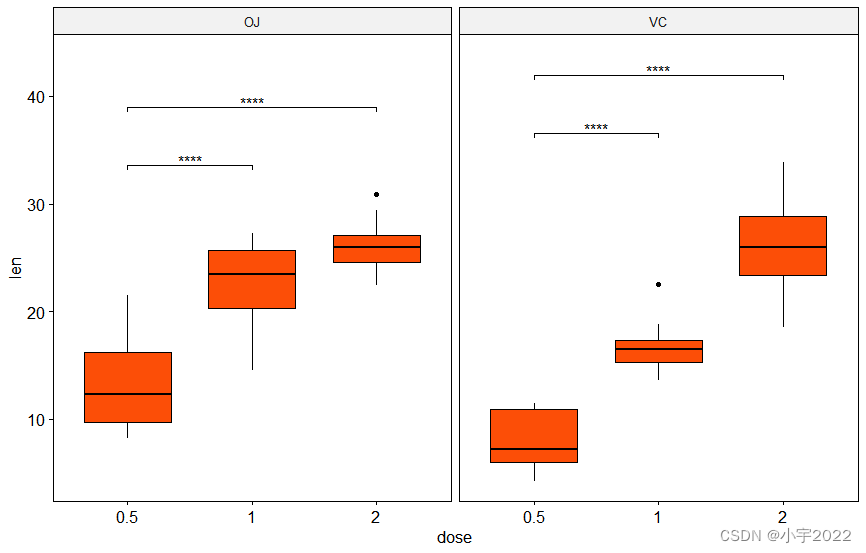
stat.test <- df %>%
group_by(supp) %>%
t_test(len ~ dose, ref.group = "0.5")
# Box plots with p-values
stat.test <- stat.test %>% add_y_position()
ggboxplot(df, x = "dose", y = "len", fill = "#FC4E07", facet.by = "supp") +
stat_pvalue_manual(stat.test, label = "p.adj.signif", tip.length = 0.01) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.1)))
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
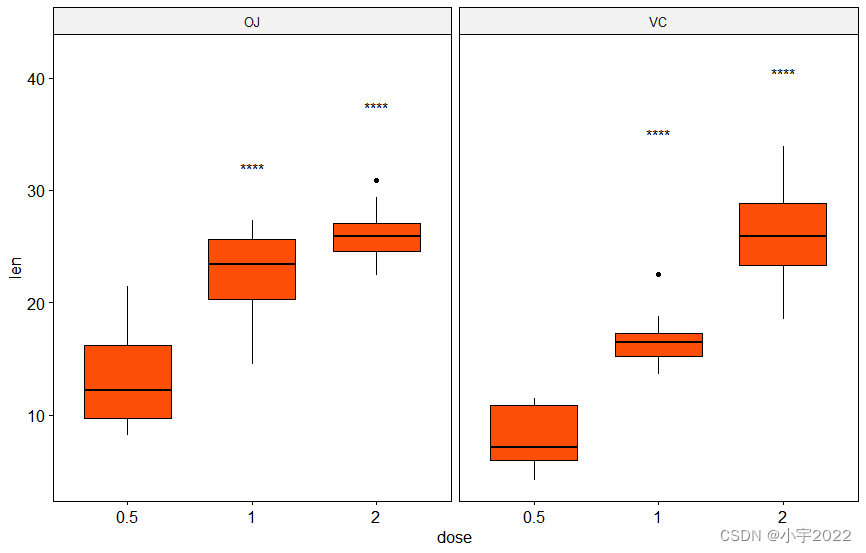
# Show only significance levels at x = group2
# Move down significance symbols using vjust
stat.test <- stat.test %>% add_y_position()
ggboxplot(df, x = "dose", y = "len", fill = "#FC4E07", facet.by = "supp") +
stat_pvalue_manual(stat.test, label = "p.adj.signif", x = "group2", vjust = 2)
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
# Bar plot with p-values
# Add 10% space on the y-axis above the box plots
stat.test <- stat.test %>% add_y_position(fun = "mean_sd")
ggbarplot(
df, x = "dose", y = "len", fill = "#FC4E07",
add = c("mean_sd", "jitter"), facet.by = "supp"
) +
stat_pvalue_manual(stat.test, label = "p.adj.signif", tip.length = 0.01) +
scale_y_continuous(expand = expansion(mult = c(0.05, 0.1)))

library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
# Create box plots with significance levels
# Hide ns (non-significant)
# Add 15% space between labels and the plot top border
stat.test <- stat.test %>% add_xy_position(x = "dose")
ggboxplot(
df, x = "dose", y = "len", fill = "#FC4E07",
facet = c("group", "supp"),
)
stat_pvalue_manual(stat.test, hide.ns = TRUE)
scale_y_continuous(expand = expansion(mult = c(0.05, 0.15)))

library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
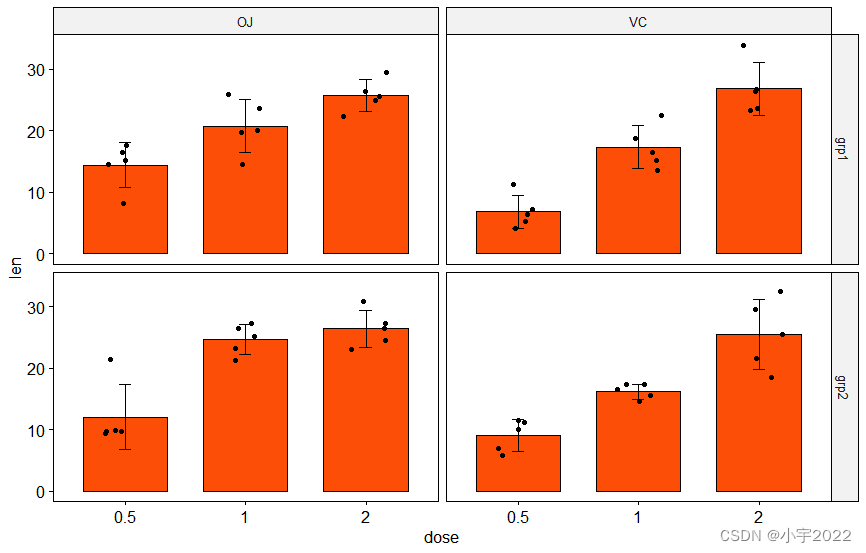
# Create bar plots with significance levels
# Hide ns (non-significant)
# Add 15% space between labels and the plot top border
stat.test <- stat.test %>% add_xy_position(x = "dose", fun = "mean_sd")
ggbarplot(
df, x = "dose", y = "len", fill = "#FC4E07",
add = c("mean_sd", "jitter"), facet = c("group", "supp")
)
stat_pvalue_manual(stat.test, hide.ns = TRUE)
scale_y_continuous(expand = expansion(mult = c(0.05, 0.15)))
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
# Box plots
bxp <- ggboxplot(
df, x = "supp", y = "len", color = "dose",
palette = "jco", facet.by = "group"
)
bxp

library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
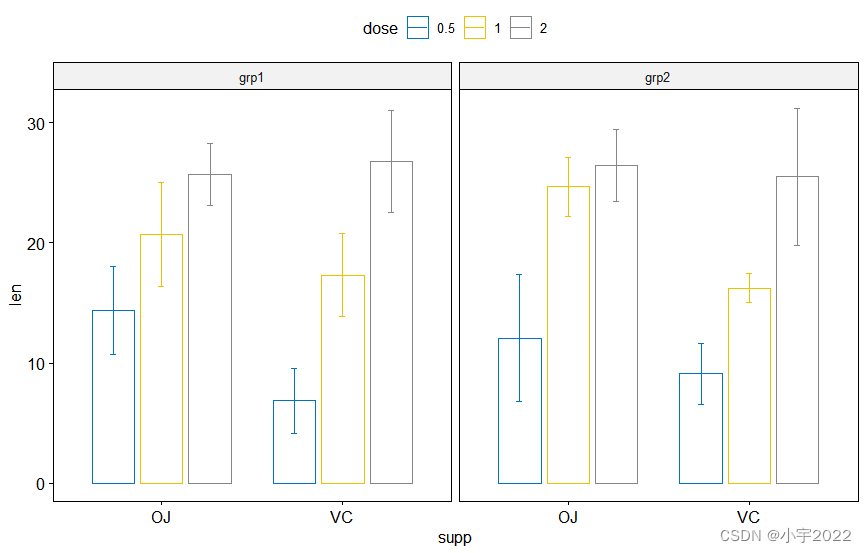
# Bar plots
bp <- ggbarplot(
df, x = "supp", y = "len", color = "dose",
palette = "jco", add = "mean_sd",
position = position_dodge(0.8),
facet.by = "group"
)
bp
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
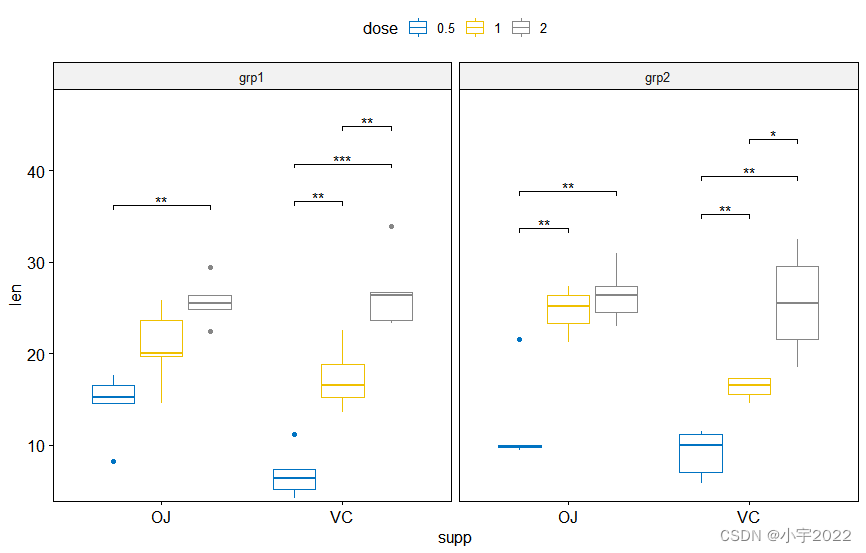
stat.test <- df %>%
group_by(supp, group) %>%
t_test(len ~ dose)
# Box plots with p-values
# Hide ns (non-significant)
stat.test <- stat.test %>%
add_xy_position(x = "supp", dodge = 0.8)
bxp +
stat_pvalue_manual(
stat.test, label = "p.adj.signif", tip.length = 0.01,
hide.ns = TRUE
) +
scale_y_continuous(expand = expansion(mult = c(0.01, 0.1)))
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
# Bar plots with p-values
stat.test <- stat.test %>%
add_xy_position(x = "supp", fun = "mean_sd", dodge = 0.8)
bp
stat_pvalue_manual(
stat.test, label = "p.adj.signif", tip.length = 0.01,
hide.ns = TRUE
)
scale_y_continuous(expand = expansion(mult = c(0.01, 0.1)))
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
stat.test <- df %>%
group_by(supp, group) %>%
t_test(len ~ dose, ref.group = "0.5")
stat.test
# Box plots with p-values
stat.test <- stat.test %>%
add_xy_position(x = "supp", dodge = 0.8)
bxp +
stat_pvalue_manual(
stat.test, label = "p.adj.signif", tip.length = 0.01
) +
scale_y_continuous(expand = expansion(mult = c(0.01, 0.1)))
library(ggpubr)
library(rstatix)
# Transform `dose` into factor variable
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
# Create a bar plot with error bars (mean +/- sd)
bp <- ggbarplot(
df, x = "supp", y = "len", add = c("mean_sd", "jitter"),
fill = "#00AFBB", facet.by = "dose"
)
stat.test <- df %>%
group_by(supp, group) %>%
t_test(len ~ dose, ref.group = "0.5")
stat.test
# Bar plots with p-values
stat.test <- stat.test %>%
add_xy_position(x = "supp", fun = "mean_sd", dodge = 0.8)
bp
stat_pvalue_manual(
stat.test, label = "p.adj.signif", tip.length = 0.01
)
scale_y_continuous(expand = expansion(mult = c(0.01, 0.1)))

边栏推荐
- HOW TO ADD P-VALUES ONTO A GROUPED GGPLOT USING THE GGPUBR R PACKAGE
- HOW TO EASILY CREATE BARPLOTS WITH ERROR BARS IN R
- GGPUBR: HOW TO ADD ADJUSTED P-VALUES TO A MULTI-PANEL GGPLOT
- GGPlot Examples Best Reference
- Beautiful and intelligent, Haval H6 supreme+ makes Yuanxiao travel safer
- K-Means Clustering Visualization in R: Step By Step Guide
- MySql存储过程游标遍历结果集
- Principle of scalable contract delegatecall
- bedtools使用教程
- 多文件程序X32dbg动态调试
猜你喜欢
![[visual studio 2019] create and import cmake project](/img/51/6c2575030c5103aee6c02bec8d5e77.jpg)
[visual studio 2019] create and import cmake project

Develop scalable contracts based on hardhat and openzeppelin (II)
ESP32音频框架 ESP-ADF 添加按键外设流程代码跟踪

念念不忘,必有回响 | 悬镜诚邀您参与OpenSCA用户有奖调研
Redis超出最大内存错误OOM command not allowed when used memory &gt; 'maxmemory'

解决uniapp列表快速滑动页面数据空白问题

FLESH-DECT(MedIA 2021)——一个material decomposition的观点

C#多维数组的属性获取方法及操作注意
mysql链表数据存储查询排序问题

How to Create a Beautiful Plots in R with Summary Statistics Labels
随机推荐
How to Easily Create Barplots with Error Bars in R
通讯录的实现(文件版本)
php 二维、多维 数组打乱顺序,PHP_php打乱数组二维数组多维数组的简单实例,php中的shuffle函数只能打乱一维
qt 仪表自定义控件
CMake交叉编译
Power Spectral Density Estimates Using FFT---MATLAB
Develop scalable contracts based on hardhat and openzeppelin (I)
K-Means Clustering Visualization in R: Step By Step Guide
The computer screen is black for no reason, and the brightness cannot be adjusted.
Summary of data export methods in powerbi
Eight sorting summaries
C#基于当前时间,获取唯一识别号(ID)的方法
解决uniapp列表快速滑动页面数据空白问题
Tidb DM alarm DM_ sync_ process_ exists_ with_ Error troubleshooting
C # method of obtaining a unique identification number (ID) based on the current time
Data analysis - Matplotlib sample code
ROS lacks catkin_ pkg
PHP 2D and multidimensional arrays are out of order, PHP_ PHP scrambles a simple example of a two-dimensional array and a multi-dimensional array. The shuffle function in PHP can only scramble one-dim
多文件程序X32dbg动态调试
预言机链上链下调研