当前位置:网站首页>Rdkit | compound library based on murcko skeleton clustering
Rdkit | compound library based on murcko skeleton clustering
2022-06-21 07:33:00 【Dazed flounder】
RDKit | be based on Murcko Skeleton clustering compound library
Assessment of compound diversity
One approach is to vectorize compounds using appropriate fingerprint techniques and evaluate the distance between them . This method is often used , But it is difficult for humans to intuitively understand the distance between compounds .
be based on Murcko skeleton , The molecules are roughly clustered in the compound skeleton , And the top compounds in each cluster are all candidate compounds . This method is very consistent with human intuition , Therefore, it can be expected that people will automatically narrow the range of compounds by looking at the list of candidate compounds .

Murcko Skeleton generation
be based on Murcko Skeleton clustering compound library
Import library
import numpy as np
from rdkit import Chem
from rdkit.Chem import Draw
from rdkit.Chem.Scaffolds import MurckoScaffold
from rdkit.Chem.Draw import IPythonConsole
Load data
sdfloader = Chem.SDMolSupplier("Enamine_Premium_processed.sdf") ### load from multisdfile
mol_list = [ mol for mol in [sdfloader[i] for i in range(len(sdfloader))] if mol is not None]
smi_scaffolds = [ MurckoScaffold.MurckoScaffoldSmiles(mol=mol, includeChirality=False) for mol in mol_list]
mol_scaffolds = [Chem.MolFromSmiles(smi_scaffold) for smi_scaffold in smi_scaffolds]
Visualize the original compound
Draw.MolsToGridImage(mol_list[:9], molsPerRow=3, subImgSize=(300,300))

Visualize skeleton compounds
Draw.MolsToGridImage(mol_scaffolds[:9], molsPerRow=3, subImgSize=(300,300))
be based on Murcko Skeleton clustering
scaffolds = {
}
clusters_list =[]
idx = 1
for mol in mol_list:
scaffold_smi = MurckoScaffold.MurckoScaffoldSmiles(mol=mol, includeChirality=False)
if scaffold_smi not in scaffolds.keys():
scaffolds[scaffold_smi] = idx
idx+=1
cluster_id = scaffolds[scaffold_smi]
clusters_list.append(cluster_id)
print("Num of dataset:",len(mol_list))
Num of dataset: 128816
print("Num of Murcko scaffolds in dataset:",len(scaffolds.keys()))
clustering 11 A cluster of , Look at its compounds
clusters_list = np.array(clusters_list)
idx_c15 = np.where(clusters_list==11)[0]
mol_list_c15 = [ mol_list[i] for i in idx_c15]
Draw.MolsToGridImage(mol_list_c15, molsPerRow=3, subImgSize=(300,300))

边栏推荐
- Unittest use
- PostgreSQL database firstborn - background first-class citizen process startupdatabase startupxlog function enters recovery mode
- Postman publishing API documentation
- Simulate long press event of mobile device
- Wechat applet_ 5. Global configuration
- CUDA or FPGA for special purpose 3D graphics computations? [closed]
- App Safety Penetration Test detailed Method Flow
- Easyexcel exclude display field-02
- [regular expression daily skill] escape characters with special meanings in regular expressions
- [telnet] telnet installation and configuration
猜你喜欢

Wechat applet_ 5. Page configuration

MATLAB快速入门

In order to thoroughly understand the problem of garbled code, I dug up the history of the character set in a rage

Wechat applet_ 3. Wxml template syntax

Hisilicon series mass production hardware commissioning record

. Net 4.5 asynchronous programming pilot (async and await)
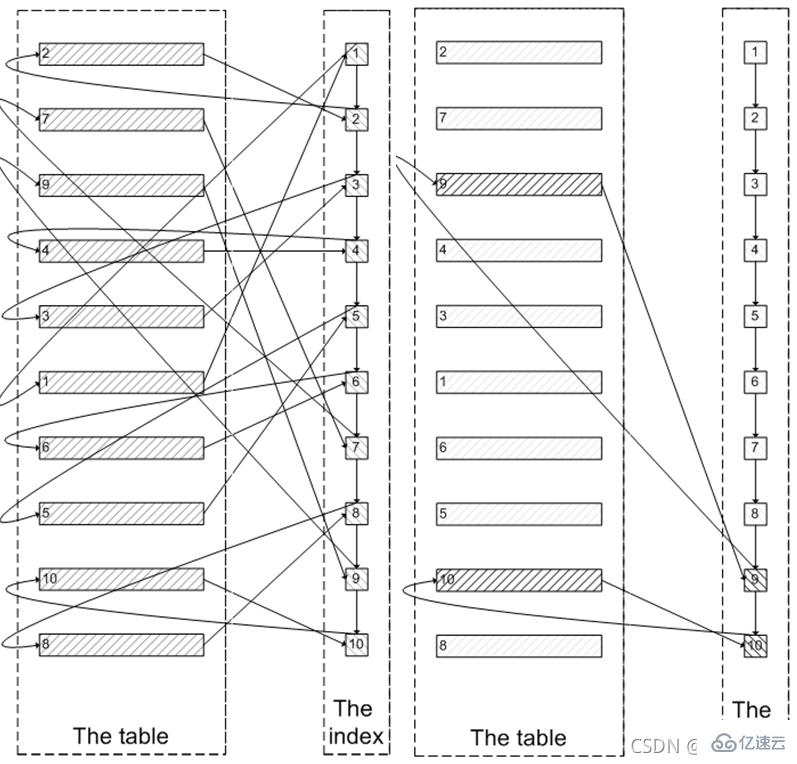
How to optimize MySQL paging query

Kubernetes pod的生命周期
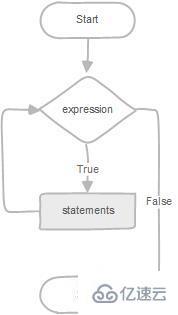
How to write circular statements in MySQL stored procedures

动态规划解决打家劫舍问题
随机推荐
23 parameter estimation -- interval estimation of a population parameter
Transport layer TCP header - serial number and acknowledgement number
EasyExcel-排除展示字段-02
C language conditional operator?: The only ternary operator
Course design of supply chain modeling and simulation based on Flexsim
Is the account with low commission safe? Is there a shortage of funds
Best practice | how to use Tencent cloud micro build to develop enterprise portal applications from 0 to 1
Tensorrt笔记(三)参考整理
[graduation season - advanced technology Er]: the technology sharing of senior college students and the future encouragement
Record context information through ThreadLocal (record user information to realize global operation)
JS operation cookie, JS setting cookie value, JS reading cookie value
Actual battle of wechat applet project -- music applet developed based on wyy music real interface
X86 CPU access DRAM and PCI
QML control type: drawer
mysql存储过程中的循环语句怎么写
RDKit | 拓扑极性表面积(TPSA)
如何让mysql不区分大小写
Exclusive Xiaoman education, medical and aesthetic education, and no direct marketing by stages
Postman publishing API documentation
[OSG] OSG development (03) -- build the osgqt Library of MSVC version