当前位置:网站首页>Work report of epidemic data analysis platform [6] visual drawing
Work report of epidemic data analysis platform [6] visual drawing
2022-06-12 04:14:00 【m0_ fifty-five million six hundred and seventy-five thousand ei】
Plotly+Dash
Using the command line pip Direct installation dash
pip install dash
Installation consists of three parts :
html、dcc、dashtable、plotly.
If you want compatibility Jupiter Related to the environment , Use :
conda install -c conda-forge -c plotly jupyter-dash
Three private open source projects , To assist in :
xinetzone/dash-xinet: simplify Dash Use .
SanstyleLab/plotly-dastsets: Store some data sets and display pictures .
xinetzone/sanstyle: Handle some common tasks .
pip install dash-xinet sanstyle
from dash_xinet.server import create_app, run_server
from dash_xinet.utils.nav import create_nav
external_stylesheets = ['https://codepen.io/chriddyp/pen/bWLwgP.css']
app = create_app(__name__, external_stylesheets=external_stylesheets)
Dash Application use and operation :
layout = ... # Layout
await run_server(app, layout, port=8050) # Start the application
About sanstyle
You can directly batch modify the file suffix :
rename_suffix(root, old, new)
root
The following directories need to be modified
old
Modified file suffix , Such as .txt
new
Modified file suffix , Such as .rst
establish Elements
from sanstyle.display.html import Embed
snippet_url = 'https://dash-tests.herokuapp.com'
figure_n_slider_dash = Embed(snippet_url + '/examples/figure-n-slider',
className='w3-pale-blue',
height=500)
figure_n_slider_dash
Import plotly-dastsets
import pandas as pd
from sanstyle.github.file import lfs_url
url = lfs_url('SanstyleLab/plotly-dastsets',
'simple/usa-agricultural-exports-2011.csv')
df = pd.read_csv(url)
df.head()
Look again plotly.
Plotly Is an interactive drawing library , Provides JavaScript、Python and R Three interfaces . We use python.
Plotly There are three different kinds of Python API, You can choose to use different methods to draw :
Object oriented API, adopt plotly.graph_objects The chart object of the module
Data driven API, By constructing something similar JSON The data structure of
Advanced drawing interface , It is the packaging of the underlying drawing method , namely plotly.express modular
object-oriented API
Use graph_objects When the module is drawing , Users need to define canvas objects go.Figure()、 Style object go.Layout()、 Trace objects such as scatter plot go.Scatter()、 Histogram go.Bar() etc. , Combine and render .
Data driven API
Plotly The visualization of is based on JSON Data structure ,trace It's a dictionary , Contains the data and colors to be drawn 、 Linear etc. drawing instructions , Used to specify how a set of data is rendered ; Organize multiple with a list trace Constitute the data, That is to say, all the things to be shown in a chart trace;layout It's also a dictionary , Used to set the layout of the chart , Including the title 、 Font and other attributes , take data and layout Together they form a chart . This method directly corresponds to Plotly Of JavaScript Implementation of the JSON API.
$ conda create -n plotly python=3.7 # Set up the name plotly Virtual environment for
$ conda activate plotly # Activate the virtual environment
$ pip install plotly # Do not specify version number
$ pip install plotly==4.14.3 # Specified version number
$ conda install -c plotly=4.14.3
$ conda install -c plotly plotly-orca==1.2.1 psutil requests
import plotly.graph_objects as go
fig = go.Figure(data=go.Bar(x=[1, 2, 3], y=[2, 3, 1]))
fig.show()

Let's look at the specific drawing steps .
Import
import warnings
import pandas as pd
import plotly.express as px
import plotly.graph_objs as go
warnings.filterwarnings('ignore')
# china_history
# Line chart of cumulative diagnosis in China
def china_confirm(df):
df0 = df[['date', 'confirm']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='confirm',
title=' Broken line chart of domestic diagnosis ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# Newly added diagnostic line chart in China
def china_confirm_add(df):
df0 = df[['date', 'confirm_add']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='confirm_add',
title=' Newly added diagnostic line chart in China ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# Domestic cumulative cure line chart
def china_heal(df):
df0 = df[['date', 'heal']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='heal',
title=' Domestic cumulative cure line chart ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# New healing line chart in China
def china_heal_add(df):
df0 = df[['date', 'heal_add']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='heal_add',
title=' New healing line chart in China ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# Line chart of cumulative domestic deaths
def china_dead(df):
df0 = df[['date', 'dead']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='dead',
title=' Line chart of cumulative domestic deaths ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# New death line chart in China
def china_dead_add(df):
df0 = df[['date', 'dead_add']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='dead_add',
title=' New death line chart in China ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# Domestic cumulative suspected line chart
def china_suspect(df):
df0 = df[['date', 'suspect']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='suspect',
title=' Domestic cumulative suspected line chart ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# Newly added suspected line chart in China
def china_suspect_add(df):
df0 = df[['date', 'suspect_add']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='suspect_add',
title=' Newly added suspected line chart in China ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# owid-covid-data
# All countries Line chart of total cases
def country_total_cases(df, country_name):
df0 = df[['date', 'total_cases']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='total_cases',
title=country_name + ' Line chart of total cases ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries New case line chart
def country_new_cases(df, country_name):
df0 = df[['date', 'new_cases']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='new_cases',
title=country_name + ' Line chart of new cases ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Line chart of total death
def country_total_deaths(df, country_name):
df0 = df[['date', 'total_deaths']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='total_deaths',
title=country_name + ' Line chart of total death ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries New death line chart
def country_new_deaths(df, country_name):
df0 = df[['date', 'new_deaths']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='new_deaths',
title=country_name + ' New death line chart ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Unit case line chart
def country_total_cases_per_million(df, country_name):
df0 = df[['date', 'total_cases_per_million']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='total_cases_per_million',
title=country_name + ' Unit case line chart ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Line chart of newly added cases in the unit
def country_new_cases_per_million(df, country_name):
df0 = df[['date', 'new_cases_per_million']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='new_cases_per_million',
title=country_name + ' Line chart of newly added cases in the unit ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Unit total death line chart
def country_total_deaths_per_million(df, country_name):
df0 = df[['date', 'total_deaths_per_million']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='total_deaths_per_million',
title=country_name + ' Unit total death line chart ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Unit new death line chart
def country_new_deaths_per_million(df, country_name):
df0 = df[['date', 'new_deaths_per_million']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='new_deaths_per_million',
title=country_name + ' Unit new death line chart ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Line graph of virus spreading rate
def country_reproduction_rate(df, country_name):
df0 = df[['date', 'reproduction_rate']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='reproduction_rate',
title=country_name + ' Line graph of virus spreading rate ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Line chart of number of severe patients
def country_icu_patients(df, country_name):
df0 = df[['date', 'icu_patients']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='icu_patients',
title=country_name + ' Line chart of number of severe patients ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Broken line chart of the number of severe patients per unit
def country_icu_patients_per_million(df, country_name):
df0 = df[['date', 'icu_patients_per_million']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='icu_patients_per_million',
title=country_name + ' Broken line chart of the number of severe patients per unit ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Line chart of nucleic acid detection quantity
def country_total_tests(df, country_name):
df0 = df[['date', 'icu_patients_per_million']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='icu_patients_per_million',
title=country_name + ' Broken line chart of the number of severe patients per unit ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries New line chart of nucleic acid detection quantity
def country_new_tests(df, country_name):
df0 = df[['date', 'new_tests']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='new_tests',
title=country_name + ' New line chart of nucleic acid detection quantity ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Line chart of unit nucleic acid detection quantity
def country_total_tests_per_thousand(df, country_name):
df0 = df[['date', 'total_tests_per_thousand']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='total_tests_per_thousand',
title=country_name + ' Line chart of unit nucleic acid detection quantity ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Line chart of new nucleic acid detection quantity per unit
def country_new_tests_per_thousand(df, country_name):
df0 = df[['date', 'new_tests_per_thousand']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='new_tests_per_thousand',
title=country_name + ' Line chart of new nucleic acid detection quantity per unit ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Positive rate line chart
def country_positive_rate(df, country_name):
df0 = df[['date', 'positive_rate']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='positive_rate',
title=country_name + ' Positive rate line chart ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Map of total vaccination amount
def country_total_vaccinations(df, country_name):
df0 = df[['date', 'total_vaccinations']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='total_vaccinations',
title=country_name + ' Map of total vaccination amount ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Number of vaccinated people
def country_people_vaccinated(df, country_name):
df0 = df[['date', 'people_vaccinated']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='people_vaccinated',
title=country_name + ' Number of vaccinated people ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries The number of people vaccinated in the whole process
def country_people_fully_vaccinated(df, country_name):
df0 = df[['date', 'people_fully_vaccinated']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='people_fully_vaccinated',
title=country_name + ' The number of people vaccinated in the whole process ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Number of new vaccinations
def country_new_vaccinations(df, country_name):
df0 = df[['date', 'new_vaccinations']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='new_vaccinations',
title=country_name + ' Number of new vaccinations ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Total vaccinations per unit
def country_total_vaccinations_per_hundred(df, country_name):
df0 = df[['date', 'total_vaccinations_per_hundred']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='total_vaccinations_per_hundred',
title=country_name + ' Total vaccinations per unit ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries Number of vaccinations per unit
def country_people_vaccinated_per_hundred(df, country_name):
df0 = df[['date', 'people_vaccinated_per_hundred']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='people_vaccinated_per_hundred',
title=country_name + ' Number of vaccinations per unit ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# All countries The number of people vaccinated in the whole process of the unit
def country_people_fully_vaccinated_per_hundred(df, country_name):
df0 = df[['date', 'people_fully_vaccinated_per_hundred']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='people_fully_vaccinated_per_hundred',
title=country_name + ' The number of people vaccinated in the whole process of the unit ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# Provinces in China
# The provinces Number of confirmed cases
def province_confirm(df, province_name):
df0 = df[['date', 'confirm']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='confirm',
title=province_name + ' Number of confirmed cases ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# The provinces The number of deaths
def province_dead(df, province_name):
df0 = df[['date', 'dead']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='dead',
title=province_name + ' The number of deaths ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# The provinces The number of people cured
def province_heal(df, province_name):
df0 = df[['date', 'heal']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='heal',
title=province_name + ' The number of people cured ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# The provinces New number of confirmed cases
def province_confirm_add(df, province_name):
df0 = df[['date', 'confirm_add']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='confirm_add',
title=province_name + ' New number of confirmed cases ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# The provinces New cured people
def province_heal_add(df, province_name):
df0 = df[['date', 'heal_add']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='heal_add',
title=province_name + ' New cured people ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
# The provinces New deaths
def province_dead_add(df, province_name):
df0 = df[['date', 'dead_add']].reset_index()
df0['date'] = df0['date'].dt.date
df0 = df0.sort_values('date', ascending=False)
fig = px.line(
data_frame=df0,
x='date',
y='dead_add',
title=province_name + ' New deaths ',
height=900
)
fig.update_layout({
# 'xaxis_dtick': 1,
'xaxis_title': ' date ',
'yaxis_title': ' Number ',
})
return fig
A little bit of code for formatting
# Date format in the original data file
format1 = '%Y-%m-%d'
format2 = '%Y/%m/%d'
# Date formatting
def date_process_df1(df):
df['date'] = pd.to_datetime(df['date'], format=format1)
return df
def date_process_df2(df):
df['date'] = pd.to_datetime(df['date'], format=format2)
return df
边栏推荐
- Absolute positioning three ways to center the box
- Community Conference | the mosn community will release version 1.0 and promote the evolution of the next generation architecture
- 动规(14)-三角形最佳路径问题
- 后续版本是否会支持代码块搜索高亮显示
- Kill session? This cross domain authentication solution is really elegant!
- 成功解决:WARNING: There was an error checking the latest version of pip.
- 关于 国产麒麟Qt编译报错“xxx.pri has modification time xxxx s in the futrue“ 的解决方法
- 电商中台系统架构
- Successfully solved: warning: there was an error checking the latest version of pip
- KV storage separation principle and performance evaluation of nebula graph
猜你喜欢

(idea)the file size(2.85M) exceeds configured limit(2.56M). Code insight features are not available

Solution en cas de défaillance du script Unity

Notes on relevant knowledge points such as original code / inverse code / complement code, size end, etc

Cloud native overview
Enterprise Architect v16

WiFi module scheme of the wireless Internet of things, esp32-s3 chip technology, helps the equipment to be intelligent
![[FPGA chaos] implementation of FPGA based chaotic system Verilog](/img/c0/cda4644168264b7531b67ef16e801a.png)
[FPGA chaos] implementation of FPGA based chaotic system Verilog

Paper recommendation: relicv2, can the new self supervised learning surpass supervised learning on RESNET?
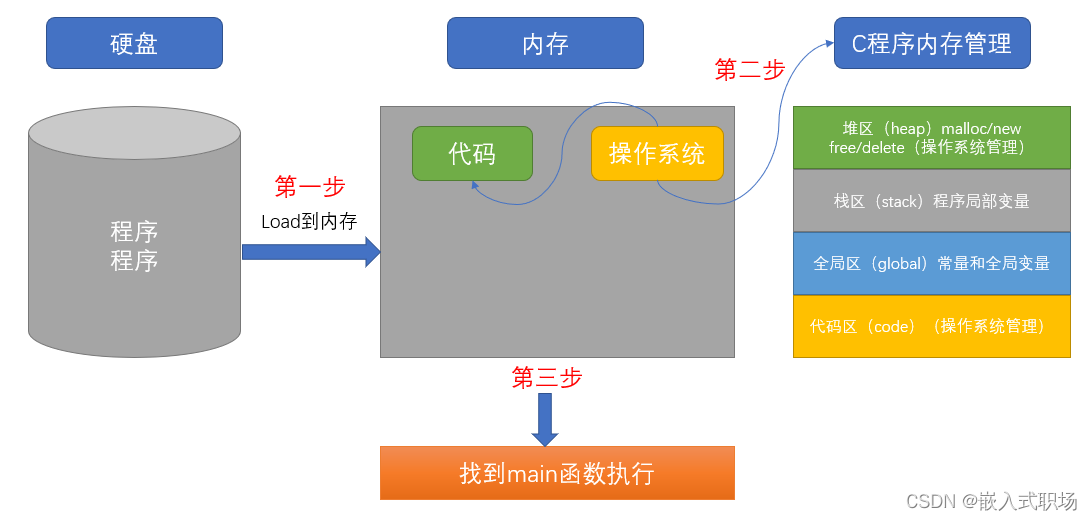
【C语言】程序的内存四区模型
What are the black box test case design methods in software testing methods?
随机推荐
MySQL create user and authorize
後續版本是否會支持代碼塊搜索高亮顯示
Evolution and practice of Unicom real-time computing platform
E-commerce middle office system architecture
Successfully solved: warning: there was an error checking the latest version of pip
成功解决:WARNING: There was an error checking the latest version of pip.
Mysql主从搭建与Django实现读写分离
分布式锁介绍
Network tester operation manual renix rack management
Kotlin 启动协程、launch 与async的区别、按照顺序启动协程
Construction case of Expressway Precast Beam Yard (with scheme text)
Call reminder
疫情数据分析平台工作报告【6】可视化绘图
Image mosaic based on transformation matrix
调用提醒事项
D1 哪吒开发板 上电记录
智能面板WiFi聯動技術,ESP32無線芯片模組,物聯網WiFi通信應用
Yyds dry inventory MySQL learning - how transactions are isolated
【C语言】封装接口(加减乘除)
Implementation of fitness club management system based on SSH