当前位置:网站首页>[data mining] task 6: DBSCAN clustering
[data mining] task 6: DBSCAN clustering
2022-07-03 01:34:00 【zstar-_】
requirement
Programming to realize DBSCAN Clustering of the following data
Data acquisition :https://download.csdn.net/download/qq1198768105/85865302
Import library and global settings
from scipy.io import loadmat
import matplotlib.pyplot as plt
import numpy as np
from sklearn.cluster import DBSCAN
from sklearn import datasets
import pandas as pd
plt.rcParams['font.sans-serif'] = ["SimHei"]
plt.rcParams["axes.unicode_minus"] = False
DBSCAN Description of clustering parameters
eps:ϵ- Distance threshold of neighborhood , The distance from the sample exceeds ϵ The sample point of is not in ϵ- In the neighborhood , The default value is 0.5.
min_samples: The minimum number of points to form a high-density area . As the core point, the neighborhood ( That is, take it as the center of the circle ,eps Is a circle of radius , Including points on the circle ) Minimum number of samples in ( Including the point itself ).
if y=-1, Is the outlier
because DBSCAN The generated category is uncertain , Therefore, define a function to filter out the most appropriate parameters that meet the specified category .
The appropriate criterion is to minimize the number of outliers
def search_best_parameter(N_clusters, X):
min_outliners = 999
best_eps = 0
best_min_samples = 0
# Iterating different eps value
for eps in np.arange(0.001, 1, 0.05):
# Iterating different min_samples value
for min_samples in range(2, 10):
dbscan = DBSCAN(eps=eps, min_samples=min_samples)
# Model fitting
y = dbscan.fit_predict(X)
# Count the number of clusters under each parameter combination (-1 Indicates an outlier )
if len(np.argwhere(y == -1)) == 0:
n_clusters = len(np.unique(y))
else:
n_clusters = len(np.unique(y)) - 1
# Number of outliers
outliners = len([i for i in y if i == -1])
if outliners < min_outliners and n_clusters == N_clusters:
min_outliners = outliners
best_eps = eps
best_min_samples = min_samples
return best_eps, best_min_samples
# Import data
colors = ['green', 'red', 'blue']
smile = loadmat('data- Density clustering /smile.mat')
smile data
X = smile['smile']
eps, min_samples = search_best_parameter(3, X)
dbscan = DBSCAN(eps=eps, min_samples=min_samples)
y = dbscan.fit_predict(X)
# Visualization of clustering results
plt.figure(figsize=(20, 15))
plt.subplot(2, 2, 1)
for i in range(len(smile['smile'])):
plt.scatter(smile['smile'][i][0], smile['smile'][i][1],
color=colors[int(smile['smile'][i][2])])
plt.title(" Raw data ")
plt.subplot(2, 2, 2)
for i in range(len(y)):
plt.scatter(smile['smile'][i][0], smile['smile'][i][1], color=colors[y[i]])
plt.title(" After clustering data ")
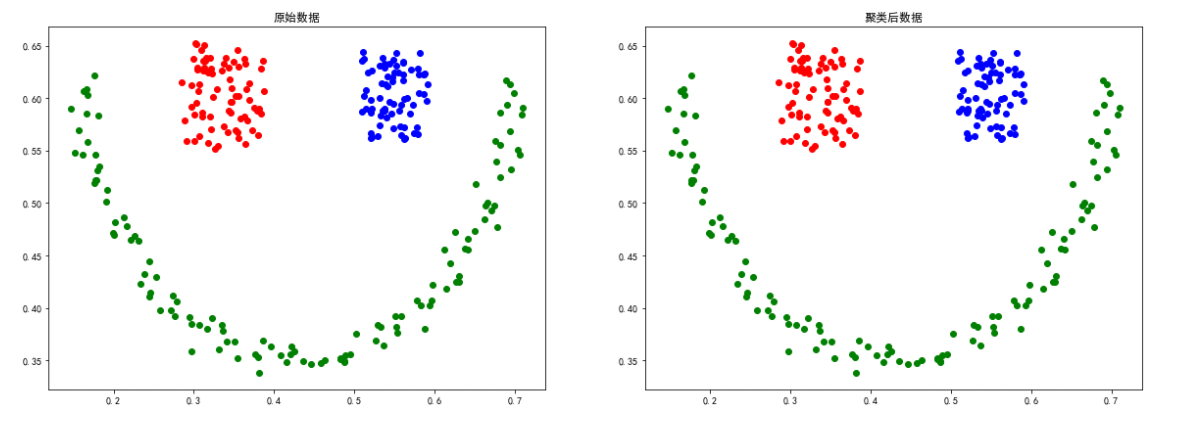
sizes5 data
# Import data
colors = ['blue', 'green', 'red', 'black', 'yellow']
sizes5 = loadmat('data- Density clustering /sizes5.mat')
X = sizes5['sizes5']
eps, min_samples = search_best_parameter(4, X)
dbscan = DBSCAN(eps=eps, min_samples=min_samples)
y = dbscan.fit_predict(X)
# Visualization of clustering results
plt.figure(figsize=(20, 15))
plt.subplot(2, 2, 1)
for i in range(len(sizes5['sizes5'])):
plt.scatter(sizes5['sizes5'][i][0], sizes5['sizes5']
[i][1], color=colors[int(sizes5['sizes5'][i][2])])
plt.title(" Raw data ")
plt.subplot(2, 2, 2)
for i in range(len(y)):
if y[i] != -1:
plt.scatter(sizes5['sizes5'][i][0], sizes5['sizes5']
[i][1], color=colors[y[i]])
plt.title(" After clustering data ")
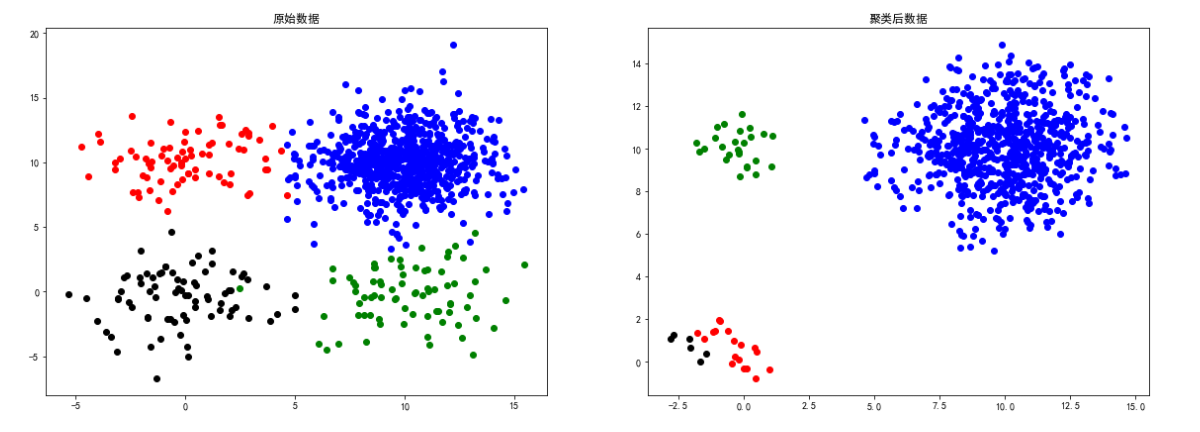
square1 data
# Import data
colors = ['green', 'red', 'blue', 'black']
square1 = loadmat('data- Density clustering /square1.mat')
X = square1['square1']
eps, min_samples = search_best_parameter(4, X)
dbscan = DBSCAN(eps=eps, min_samples=min_samples)
y = dbscan.fit_predict(X)
# Visualization of clustering results
plt.figure(figsize=(20, 15))
plt.subplot(2, 2, 1)
for i in range(len(square1['square1'])):
plt.scatter(square1['square1'][i][0], square1['square1']
[i][1], color=colors[int(square1['square1'][i][2])])
plt.title(" Raw data ")
plt.subplot(2, 2, 2)
for i in range(len(y)):
plt.scatter(square1['square1'][i][0], square1['square1']
[i][1], color=colors[y[i]])
plt.title(" After clustering data ")

square4 data
# Import data
colors = ['blue', 'green', 'red', 'black',
'yellow', 'brown', 'orange', 'purple']
square4 = loadmat('data- Density clustering /square4.mat')
X = square4['b']
eps, min_samples = search_best_parameter(5, X)
dbscan = DBSCAN(eps=eps, min_samples=min_samples)
y = dbscan.fit_predict(X)
# Visualization of clustering results
plt.figure(figsize=(20, 15))
plt.subplot(2, 2, 1)
for i in range(len(square4['b'])):
plt.scatter(square4['b'][i][0], square4['b']
[i][1], color=colors[int(square4['b'][i][2])])
plt.title(" Raw data ")
plt.subplot(2, 2, 2)
for i in range(len(y)):
plt.scatter(square4['b'][i][0], square4['b']
[i][1], color=colors[y[i]])
plt.title(" After clustering data ")
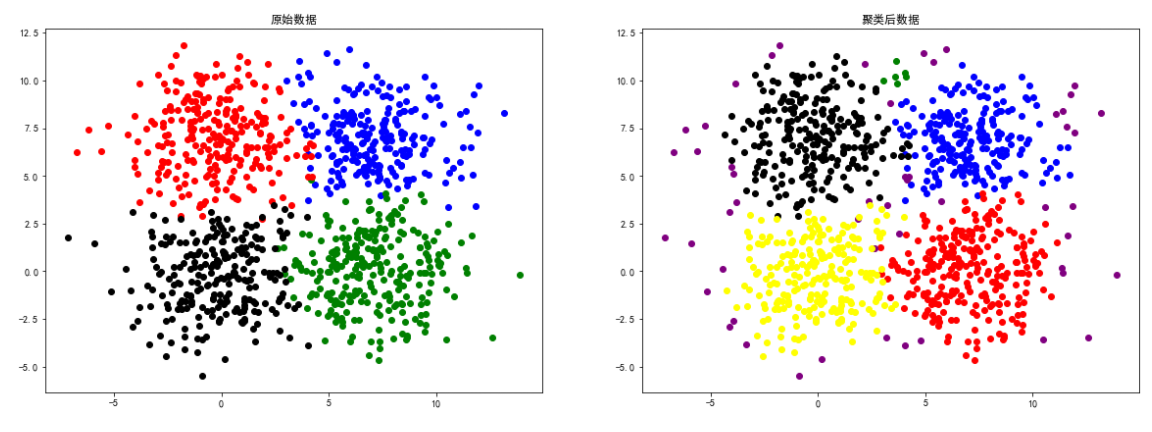
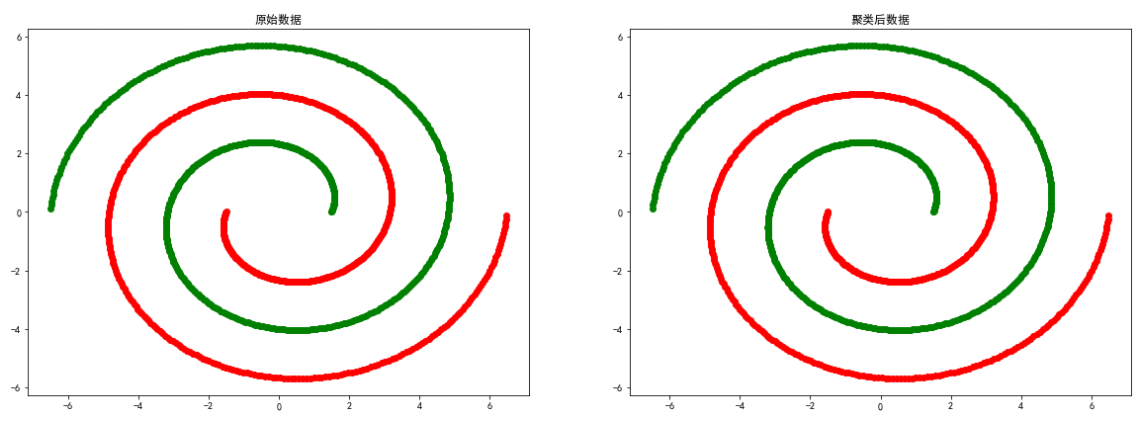
spiral data
# Import data
colors = ['green', 'red']
spiral = loadmat('data- Density clustering /spiral.mat')
X = spiral['spiral']
eps, min_samples = search_best_parameter(2, X)
dbscan = DBSCAN(eps=eps, min_samples=min_samples)
y = dbscan.fit_predict(X)
# Visualization of clustering results
plt.figure(figsize=(20, 15))
plt.subplot(2, 2, 1)
for i in range(len(spiral['spiral'])):
plt.scatter(spiral['spiral'][i][0], spiral['spiral']
[i][1], color=colors[int(spiral['spiral'][i][2])])
plt.title(" Raw data ")
plt.subplot(2, 2, 2)
for i in range(len(y)):
plt.scatter(spiral['spiral'][i][0], spiral['spiral']
[i][1], color=colors[y[i]])
plt.title(" After clustering data ")
moon data
# Import data
colors = ['green', 'red']
moon = loadmat('data- Density clustering /moon.mat')
X = moon['a']
eps, min_samples = search_best_parameter(2, X)
dbscan = DBSCAN(eps=eps, min_samples=min_samples)
y = dbscan.fit_predict(X)
# Visualization of clustering results
plt.figure(figsize=(20, 15))
plt.subplot(2, 2, 1)
for i in range(len(moon['a'])):
plt.scatter(moon['a'][i][0], moon['a']
[i][1], color=colors[int(moon['a'][i][2])])
plt.title(" Raw data ")
plt.subplot(2, 2, 2)
for i in range(len(y)):
plt.scatter(moon['a'][i][0], moon['a']
[i][1], color=colors[y[i]])
plt.title(" After clustering data ")

long data
# Import data
colors = ['green', 'red']
long = loadmat('data- Density clustering /long.mat')
X = long['long1']
eps, min_samples = search_best_parameter(2, X)
dbscan = DBSCAN(eps=eps, min_samples=min_samples)
y = dbscan.fit_predict(X)
# Visualization of clustering results
plt.figure(figsize=(20, 15))
plt.subplot(2, 2, 1)
for i in range(len(long['long1'])):
plt.scatter(long['long1'][i][0], long['long1']
[i][1], color=colors[int(long['long1'][i][2])])
plt.title(" Raw data ")
plt.subplot(2, 2, 2)
for i in range(len(y)):
plt.scatter(long['long1'][i][0], long['long1']
[i][1], color=colors[y[i]])
plt.title(" After clustering data ")

2d4c data
# Import data
colors = ['green', 'red', 'blue', 'black']
d4c = loadmat('data- Density clustering /2d4c.mat')
X = d4c['a']
eps, min_samples = search_best_parameter(4, X)
dbscan = DBSCAN(eps=eps, min_samples=min_samples)
y = dbscan.fit_predict(X)
# Visualization of clustering results
plt.figure(figsize=(20, 15))
plt.subplot(2, 2, 1)
for i in range(len(d4c['a'])):
plt.scatter(d4c['a'][i][0], d4c['a']
[i][1], color=colors[int(d4c['a'][i][2])])
plt.title(" Raw data ")
plt.subplot(2, 2, 2)
for i in range(len(y)):
plt.scatter(d4c['a'][i][0], d4c['a']
[i][1], color=colors[y[i]])
plt.title(" After clustering data ")

summary
The above experiment proves that DBSCAN Clustering methods are more dependent on the relevance of data points , about smile、spiral The clustering effect of equally distributed data is better .
边栏推荐
- Steps to obtain SSL certificate private key private key file
- Now that the teenager has returned, the world's fireworks are the most soothing and ordinary people return to work~
- 【数据挖掘】任务4:20Newsgroups聚类
- 一比特苦逼程序員的找工作經曆
- 【C语言】指针与数组笔试题详解
- [QT] encapsulation of custom controls
- Mathematical knowledge: divisible number inclusion exclusion principle
- d. LDC build shared library
- High-Resolution Network (篇一):原理刨析
- 【数据挖掘】任务5:K-means/DBSCAN聚类:双层正方形
猜你喜欢
看完这篇 教你玩转渗透测试靶机Vulnhub——DriftingBlues-9

MySQL - database query - condition query
![[Androd] Gradle 使用技巧之模块依赖替换](/img/5f/968db696932f155a8c4a45f67135ac.png)
[Androd] Gradle 使用技巧之模块依赖替换
![[Arduino experiment 17 L298N motor drive module]](/img/e2/4511eaa942e4a64c8ca2ee70162785.jpg)
[Arduino experiment 17 L298N motor drive module]
电信客户流失预测挑战赛
给你一个可能存在 重复 元素值的数组 numbers ,它原来是一个升序排列的数组,并按上述情形进行了一次旋转。请返回旋转数组的最小元素。【剑指Offer】

并发编程的三大核心问题 -《深入理解高并发编程》

How is the mask effect achieved in the LPL ban/pick selection stage?
Wireshark data analysis and forensics a.pacapng

MySQL foundation 04 MySQL architecture
随机推荐
Steps to obtain SSL certificate private key private key file
[day 29] given an integer, please find its factor number
一位苦逼程序员的找工作经历
Test shift right: Elk practice of online quality monitoring
对非ts/js文件模块进行类型扩充
[interview question] 1369 when can't I use arrow function?
leetcode 2097 — 合法重新排列数对
C application interface development foundation - form control (4) - selection control
Vim 9.0正式发布!新版脚本执行速度最高提升100倍
High resolution network (Part 1): Principle Analysis
Androd gradle's substitution of its use module dependency
Uniapp component -uni notice bar notice bar
Mathematical knowledge: Nim game game theory
Work experience of a hard pressed programmer
dotConnect for PostgreSQL数据提供程序
按键精灵打怪学习-自动回城路线的判断
wirehark数据分析与取证A.pacapng
Database SQL language 02 connection query
【C语言】指针与数组笔试题详解
Kivy tutorial - example of using Matplotlib in Kivy app