当前位置:网站首页>30 minutes to understand PCA principal component analysis
30 minutes to understand PCA principal component analysis
2022-07-06 18:13:00 【The way of Python data】
source : Algorithm food house
This article shares with you PCA The concept of principal component analysis and in Python The use of . I also shared two articles before , It's also very good , It can be combined to see , Deepen the understanding .
Article to read PCA The mathematical principle of the algorithm
Let's talk about the dimensionality reduction algorithm :PCA Principal component analysis
PCA Principal component analysis algorithm (Principal Components Analysis) It is one of the most commonly used dimensionality reduction algorithms . With low information loss ( Measured by the variance of distribution between samples ) Reduce the number of features .
PCA Algorithms can help Analyze the components with the greatest distribution difference in the sample ( The principal components ), Help data visualization ( Down to 2 Dimension or 3 After dimension, it can be visualized with scatter diagram ), Sometimes it can also play Reduce the noise in the sample The role of ( Part of the lost information is noise ).
One 、PCA Intuitive understanding of algorithms
Intuitively ,PCA Principal component analysis is similar to finding the direction long axis with the greatest difference in each position of a group of sample points in turn .
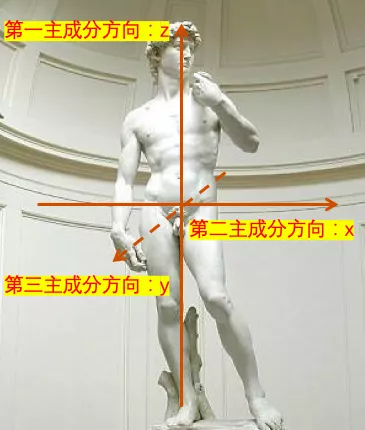
Suppose that all cells in a person are regarded as sample points one by one . These sample points can be used 3 Coordinates to represent , From left to right x Direction , From front to back y Direction , From bottom to top is z Direction .
So what is their first principal component ? The long axis corresponding to the first principal component is along the direction from the foot to the head , That is, the usual up and down direction , namely z Direction . This direction is the main long axis . The position difference of these sample points is basically 70% The above comes from the difference in this direction .
What is their second principal component ? The second principal component corresponds to the direction from left arm to right arm , That is, the usual left-right direction , namely y Direction . This direction is perpendicular to the long axis of the first principal component , The location differences of these sample points are about 20% Left and right come from the difference in this direction .
What is their third principal component ? The third principal component direction is along the direction from the chest to the back , That is, the usual fore-and-aft direction , namely x Direction . This direction is perpendicular to the long axis of the first two principal components , The position difference of sample points is generally less than 10% The difference from this direction , Of course , Except for some students who are more rounded .
Now? , If you want to reduce the coordinates of these sample points to 2 One to indicate , And keep as much information about the location differences between sample points as possible , that , obviously , The coordinates of the first principal component and the second principal component should be retained .
Now? , Suppose this person is lying on a reclining chair , Now the first principal component of these sample points is obviously no longer from bottom to top z Direction . We should rotate our coordinate system , Give Way z The axis is in the same direction as the reclining chair , This new z The direction is the first principal component direction of these sample points . Similarly , You also need to rotate x Axis and y The axis obtains the new second principal component direction and the third principal component direction .
The process of rotating the old coordinate system to find the direction of the principal component is PCA Principal component analysis .
Two 、PCA Mathematical description of the algorithm
PCA Principal component analysis (Principal Components Analysis) It's a kind of passing Orthogonal linear combination The way , Maximize the variance between samples Of Dimension reduction Method .
From a geometric point of view ,PCA The principal component analysis method can be regarded as through Orthogonal transformation , Rotate and translate the coordinate system , The first several new coordinates with the largest variance of the projection coordinates of the sample points are retained .
Here are a few key words to explain :
Dimension reduction : Take the original sample m Victor requisitioned less k Features replace . Dimension reduction algorithm can be understood as a data compression method , It may lose some information .
Orthogonal linear combination :k A new feature is through m It is produced by linear combination of Gejiu features , also k The coefficient vector of a linear combination is the unit vector , And they are orthogonal to each other .
Maximize the variance between samples : The first 1 Principal component features maximize the characteristic variance between samples , The first 2 The three principal component features are satisfying the 1 Maximize the characteristic variance between samples under the orthogonal constraints of principal components ……
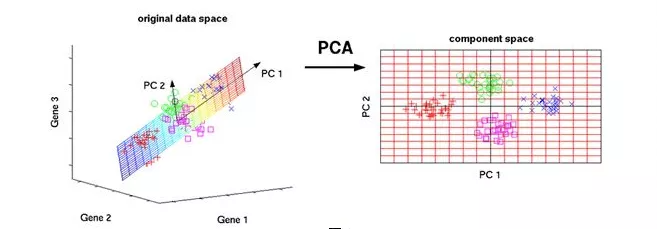
3、 ... and 、PCA Example of algorithm switching
In the following example, we call sklearn Medium PCA Dimension reduction algorithm interface , Dimensionality reduction of Boston house price data set (13 Dimension down to 7 dimension ).
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import datasets
boston = datasets.load_boston()
dfdata = pd.DataFrame(boston.data,columns = boston.feature_names)
dfdata.head()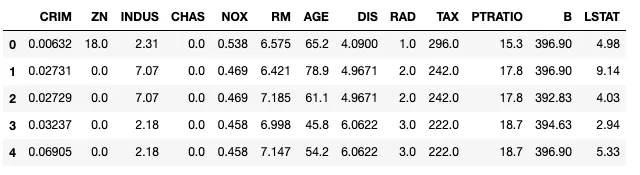
# The value range of different features varies greatly , We first normalize the standard normal
# The normalized result is taken as PCA Dimension reduction input
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(dfdata.values)
X_input = scaler.transform(dfdata.values)
# Our inputs are 506 Samples ,13 Whitman's sign
print(X_input.shape)(506, 13)# application PCA Carry out dimension reduction
from sklearn.decomposition import PCA
pca = PCA(n_components=7)
pca.fit(X_input)
X_output = pca.transform(X_input)
# After the dimension reduction , Only 7 Whitman's sign
print(X_output.shape)(506, 7)# Check the variance corresponding to each principal component and the proportion in the total variance
# You can see before 7 Principal components have explained the distribution of samples 90% The difference .
print("explained_variance:")
print(pca.explained_variance_)
print("explained_variance_ratio:")
print(pca.explained_variance_ratio_)
print("total explained variance ratio of first 7 principal components:")
print(sum(pca.explained_variance_ratio_)explained_variance:
[6.1389812 1.43611329 1.2450773 0.85927328 0.83646904 0.65870897
0.5364162 ]
explained_variance_ratio:
[0.47129606 0.11025193 0.0955859 0.06596732 0.06421661 0.05056978
0.04118124]
total explained variance ratio of first 7 principal components:
0.8990688406240493# Visualize the variance of each principal component contribution
%matplotlib inline
%config InlineBackend.figure_format = 'svg'
import matplotlib.pyplot as plt
plt.figure()
plt.plot(np.arange(1,8),pca.explained_variance_,linewidth=2)
plt.xlabel('components_n', fontsize=16)
plt.ylabel('explained_variance_', fontsize=16)
plt.show()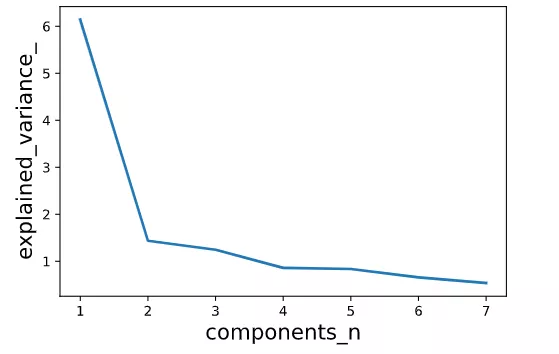
# Check the orthogonal transformation matrix corresponding to the dimension reduction , That is, each projection vector
W = pca.components_
# Verify orthogonality
np.round(np.dot(W,np.transpose(W)),6)array([[ 1., 0., -0., -0., -0., 0., -0.],
[ 0., 1., -0., 0., 0., -0., -0.],
[-0., -0., 1., 0., -0., 0., -0.],
[-0., 0., 0., 1., -0., -0., 0.],
[-0., 0., -0., -0., 1., 0., 0.],
[ 0., -0., 0., -0., 0., 1., -0.],
[-0., -0., -0., 0., 0., -0., 1.]])# Visualize the first two dimensions of the reduced dimension data
%matplotlib inline
%config InlineBackend.figure_format = 'svg'
plt.scatter(X_output[:,0],X_output[:,1])
plt.xlabel("components_1")
plt.ylabel("components_2")
plt.show()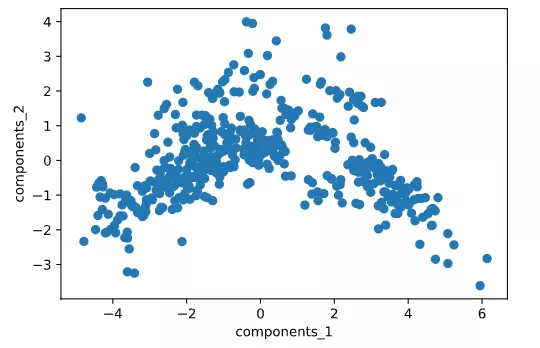
Four 、PCA The mathematical principle of the algorithm
High number reminder ahead .
Underneath PCA Deduction of mathematical principle of Algorithm , Certificate No k The principal component projection vector is exactly the... Of the covariance matrix of the sample k Eigenvectors corresponding to large eigenvalues .
The following deduction will use some knowledge of linear algebra and calculus in Advanced Mathematics .
Students without relevant mathematical foundation can skip , In practice, just master PCA Intuitive concept of the algorithm and how to use it , Basically enough .
Suppose that the characteristics of the sample are represented by a matrix , Each line represents a sample , Each column represents a feature .
Assuming that Dimensional , I.e. samples , Whitman's sign .
Now look for the first principal component .
Suppose the first principal component projection vector is , It is a unit column vector , Dimension is ,
Then the coordinate after projection is
The coordinate variance between the projected samples can be expressed as
Bars mean averaging the samples , Because it has nothing to do with the sample , It can be reduced to :
be aware It happens to be The covariance matrix of .
Next we want to maximize the coordinate variance , At the same time, the unit length constraint must be satisfied :
For extreme value conditions with constraints , We can apply Lagrange multiplier method in calculus . Construct the following Lagrange function .
402 Payment Required
The subscript representation can be used to prove , For quadratic form , There is the following vector derivation formula :
We can deduce that :
402 Payment Required
It can be seen that , Is the eigenvalue of the covariance matrix , Is the corresponding eigenvector . Because the covariance matrix is a real symmetric matrix , Its characteristic value must be greater than or equal to 0.
meanwhile , The coordinate variance between the projected samples can be expressed as :
402 Payment Required
To maximize the coordinate variance between samples , The largest eigenvalue of the covariance matrix should be taken , Then it should be the eigenvector corresponding to the maximum eigenvalue of the covariance matrix .
Similarly , It can be proved that k The first principal component projection vector is the... Of the covariance matrix k Eigenvectors corresponding to large eigenvalues .
Because the eigenvectors of different eigenvalues of the matrix are orthogonal to each other , Therefore, these projection vectors satisfy the orthogonal condition .
边栏推荐
- 第三季百度网盘AI大赛盛夏来袭,寻找热爱AI的你!
- F200 - UAV equipped with domestic open source flight control system based on Model Design
- OliveTin能在网页上安全运行shell命令(上)
- C language exchanges two numbers through pointers
- 【剑指 Offer】 60. n个骰子的点数
- Principle and usage of extern
- UDP协议:因性善而简单,难免碰到“城会玩”
- std::true_type和std::false_type
- Release of the sample chapter of "uncover the secrets of asp.net core 6 framework" [200 pages /5 chapters]
- 带你穿越古罗马,元宇宙巴士来啦 #Invisible Cities
猜你喜欢
kivy教程之在 Kivy 中支持中文以构建跨平台应用程序(教程含源码)
node の SQLite
Declval (example of return value of guidance function)

Why should Li Shufu personally take charge of building mobile phones?
趣-关于undefined的问题

Getting started with pytest ----- test case pre post, firmware
Appium automated test scroll and drag_ and_ Drop slides according to element position
ASEMI整流桥DB207的导通时间与参数选择

2019阿里集群数据集使用总结

Interesting - questions about undefined
随机推荐
Take you through ancient Rome, the meta universe bus is coming # Invisible Cities
In terms of byte measurement with an annual salary of 30W, automated testing can be learned in this way
SQL statement optimization, order by desc speed optimization
C语言通过指针交换两个数
【剑指 Offer】 60. n个骰子的点数
微信为什么使用 SQLite 保存聊天记录?
RB157-ASEMI整流桥RB157
scratch疫情隔离和核酸检测模拟 电子学会图形化编程scratch等级考试三级真题和答案解析2022年6月
MSF horizontal MSF port forwarding + routing table +socks5+proxychains
Jerry's watch deletes the existing dial file [chapter]
Awk command exercise
Alertmanager sends the alarm email and specifies it as the Alibaba mailbox of the company
Video fusion cloud platform easycvr adds multi-level grouping, which can flexibly manage access devices
Running the service with systemctl in the container reports an error: failed to get D-Bus connection: operation not permitted (solution)
2019阿里集群数据集使用总结
Getting started with pytest ----- allow generate report
OliveTin能在网页上安全运行shell命令(上)
Transport layer congestion control - slow start and congestion avoidance, fast retransmission, fast recovery
Selected technical experts from China Mobile, ant, SF, and Xingsheng will show you the guarantee of architecture stability
容器里用systemctl运行服务报错:Failed to get D-Bus connection: Operation not permitted(解决方法)