当前位置:网站首页>[Taichi] change pbf2d (position based fluid simulation) of Taiji to pbf3d with minimal modification
[Taichi] change pbf2d (position based fluid simulation) of Taiji to pbf3d with minimal modification
2022-07-04 23:07:00 【beidou111】
Effect display 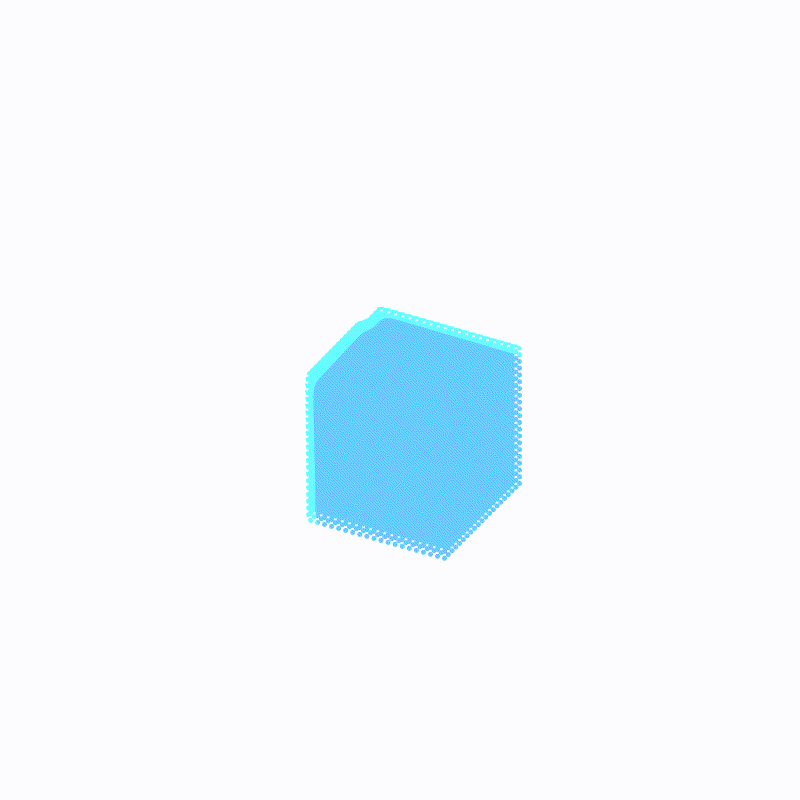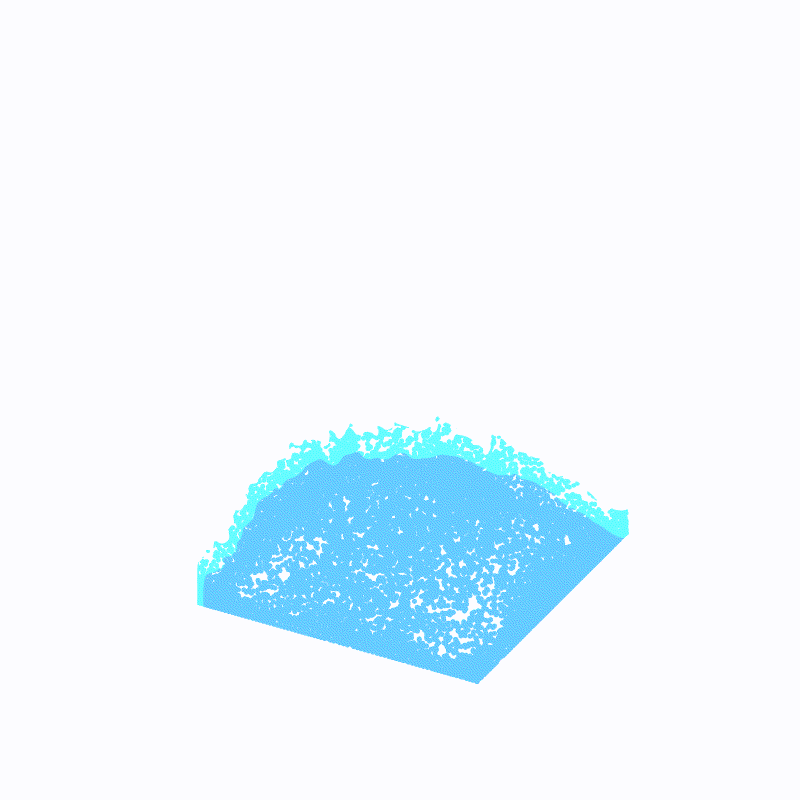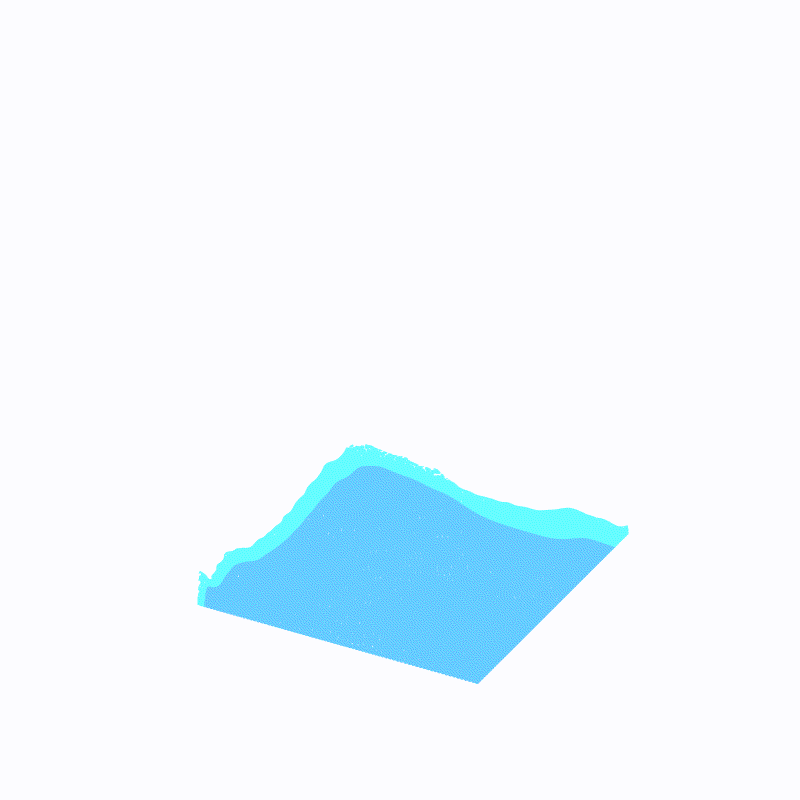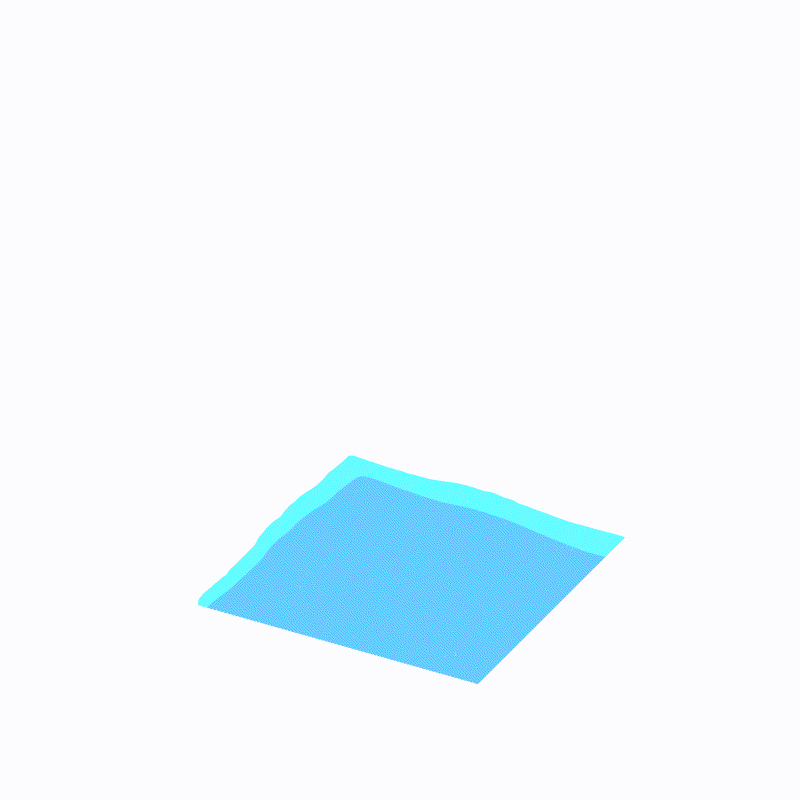
( The outermost light blue is ffmpeg Caused by the , Please ignore )
github Warehouse
https://github.com/chunleili/pbf3d
Principle explanation
https://www.bilibili.com/video/BV1qa411X7Uo/
The code itself is written very dimensionally independent . When changing, you only need to pay attention to the following points :
The technology is summarized as follows
3d Of , Either use GGUI( Reference resources mpm3d_ggui That example ), Or use 2d Rendering 3d( Reference resources mpm3d That example ). So-called 2d Rendering 3d It's actually doing it by yourself MVP Transformation ( Model 、 View 、 Projection transformation ), This point is copied directly mpm3d In the example T Function to 3d The particle coordinates of are projected to 2d On the screen .
All appear ti.Vector([0.0, 0.0]) Obviously, it should be changed into ti.Vector([0.0, 0.0, 0.0]). Empathy ti.grouped(ti.ndrange((-1, 2), (-1, 2),(-1, 2))) This is changed into 3 Dimensional . This is all because of Tai Chi ti.Vector And the trouble caused by the current inability to achieve dimension independence .
Pay attention to the data structure grid_num_particles and grid2particles These two data structures . because SNode Cannot create temporarily 4 Dimension group ( because grid_size Itself is 3 Dimensional , Then the array of particles in the mesh is the fourth dimension ), So just use ti.field Appoint shape The way
grid_num_particles It's simple , Just put ti.ij Change it to ti.ijk That's it
grid_snode = ti.root.dense(ti.ijk, grid_size)
grid_snode.place(grid_num_particles)
grid2particles = ti.field(int, (grid_size + (max_num_particles_per_cell,)))
Notice the comma here . because python When parentheses wrap tuples , Tuples can be spliced , But first int Convert to tuple , So I have to add a comma to indicate that it is not a number , It's a tuple .
The initial arrangement takes some careful thought . But it's not too hard . Refer to my last post . Or look here http://t.csdn.cn/Kpku8. In fact, that is “ Sit in rows , Eat fruit , Row after row , One floor after another ”. I didn't modify it directly k-ye Of init_particles, Instead, I just wrote a new one init function .
Some unnecessary functions can be cleaned out , such as move_board And related codes and variables . This doesn't work , Just added a fancy Mobile board .
Pay attention to adjusting the size of the world . I mean T Last in the function return Original +0.5, I changed it here to 25. Why should we change it ? because mpm3d The size of the world is 1.01.01.0 Of . and k-ye The code world size of is boundary( Let's just say boundary It's the sky box ). I've finished changing here boundary Its size is (40,40,40). You may also find mine gui.circles(T(pos)/100.0, radius=3, color=0x66ccff) This is divided by onehundred , This is also to scale the size of the world .
My advice is : The size of the world follows Kuang Ye's setting , instead of mpm3d Set up . If you follow mpm3d Set up , Nuclear radius h It will be smaller than 1, This will lead to a result : stay evaluate Kernel function , It will lead to abnormally large kernel function values . Because in spiky kernel in h Of 6 The power is the denominator !
- Last suggestion : When modifying the code , One principle of assurance is to minimize modification . The so-called can not change , I don't know whether it should be changed or not , We must change it all at once . Remember what you have changed , No change . Don't modify the shotgun style . Uncertain functions or features will be tested separately . Carefully use new features that you don't particularly understand .( Make good use of git)
The code is as follows
# Macklin, M. and Müller, M., 2013. Position based fluids. ACM Transactions on Graphics (TOG), 32(4), p.104.
# Taichi implementation by Ye Kuang (k-ye)
# Modified by Chunlei Li, change to 3D, 2022/7/4
import math
import numpy as np
import taichi as ti
ti.init(arch=ti.gpu)
screen_res = (800, 800)
screen_to_world_ratio = 20.0
boundary = (screen_res[0] / screen_to_world_ratio,
screen_res[1] / screen_to_world_ratio,
screen_res[0] / screen_to_world_ratio,)
cell_size = 2.51
cell_recpr = 1.0 / cell_size
def round_up(f, s):
return (math.floor(f * cell_recpr / s) + 1) * s
grid_size = (round_up(boundary[0], 1), round_up(boundary[1], 1), round_up(boundary[2], 1))
dim = 3
bg_color = 0x112f41
particle_color = 0x068587
boundary_color = 0xebaca2
num_particles_x = 10
# num_particles = num_particles_x * num_particles_x * 10
num_particles = 10000
max_num_particles_per_cell = 100
max_num_neighbors = 100
time_delta = 1.0 / 20.0
epsilon = 1e-5
particle_radius = 3.0
particle_radius_in_world = particle_radius / screen_to_world_ratio
# PBF params
h = 1.1
mass = 1.0
rho0 = 1.0
lambda_epsilon = 100.0
pbf_num_iters = 5
corr_deltaQ_coeff = 0.3
corrK = 0.001
# Need ti.pow()
# corrN = 4.0
neighbor_radius = h * 1.05
poly6_factor = 315.0 / 64.0 / math.pi
spiky_grad_factor = -45.0 / math.pi
old_positions = ti.Vector.field(dim, float)
positions = ti.Vector.field(dim, float)
velocities = ti.Vector.field(dim, float)
grid_num_particles = ti.field(int)
# grid2particles = ti.field(int)
particle_num_neighbors = ti.field(int)
particle_neighbors = ti.field(int)
lambdas = ti.field(float)
position_deltas = ti.Vector.field(dim, float)
ti.root.dense(ti.i, num_particles).place(old_positions, positions, velocities)
grid_snode = ti.root.dense(ti.ijk, grid_size)
grid_snode.place(grid_num_particles)
# grid_snode.dense(ti.i, max_num_particles_per_cell).place(grid2particles) #this way cannot place a 4 dimension array
grid2particles = ti.field(int, (grid_size + (max_num_particles_per_cell,)))
nb_node = ti.root.dense(ti.i, num_particles)
nb_node.place(particle_num_neighbors)
nb_node.dense(ti.j, max_num_neighbors).place(particle_neighbors)
ti.root.dense(ti.i, num_particles).place(lambdas, position_deltas)
@ti.func
def poly6_value(s, h):
result = 0.0
if 0 < s and s < h:
x = (h * h - s * s) / (h * h * h)
result = poly6_factor * x * x * x
return result
@ti.func
def spiky_gradient(r, h):
result = ti.Vector([0.0, 0.0, 0.0])
r_len = r.norm()
if 0 < r_len and r_len < h:
x = (h - r_len) / (h * h * h)
g_factor = spiky_grad_factor * x * x
result = r * g_factor / r_len
return result
@ti.func
def compute_scorr(pos_ji):
# Eq (13)
x = poly6_value(pos_ji.norm(), h) / poly6_value(corr_deltaQ_coeff * h, h)
# pow(x, 4)
x = x * x
x = x * x
return (-corrK) * x
@ti.func
def get_cell(pos):
return int(pos * cell_recpr)
@ti.func
def is_in_grid(c):
# @c: Vector(i32)
return 0 <= c[0] and c[0] < grid_size[0] and 0 <= c[1] and c[
1] < grid_size[1]
@ti.func
def confine_position_to_boundary(p):
bmin = particle_radius_in_world
bmax = ti.Vector([boundary[0], boundary[1], boundary[2]
]) - particle_radius_in_world
for i in ti.static(range(dim)):
# Use randomness to prevent particles from sticking into each other after clamping
if p[i] <= bmin:
p[i] = bmin + epsilon * ti.random()
elif bmax[i] <= p[i]:
p[i] = bmax[i] - epsilon * ti.random()
return p
@ti.kernel
def prologue():
# save old positions
for i in positions:
old_positions[i] = positions[i]
# apply gravity within boundary
for i in positions:
g = ti.Vector([0.0, -9.8, 0.0])
pos, vel = positions[i], velocities[i]
vel += g * time_delta
pos += vel * time_delta
positions[i] = confine_position_to_boundary(pos)
# clear neighbor lookup table
for I in ti.grouped(grid_num_particles):
grid_num_particles[I] = 0
for I in ti.grouped(particle_neighbors):
particle_neighbors[I] = -1
# update grid
for p_i in positions:
cell = get_cell(positions[p_i])
# ti.Vector doesn't seem to support unpacking yet
# but we can directly use int Vectors as indices
offs = ti.atomic_add(grid_num_particles[cell], 1)
grid2particles[cell, offs] = p_i
# find particle neighbors
for p_i in positions:
pos_i = positions[p_i]
cell = get_cell(pos_i)
nb_i = 0
for offs in ti.static(ti.grouped(ti.ndrange((-1, 2), (-1, 2),(-1, 2)))):
cell_to_check = cell + offs
if is_in_grid(cell_to_check):
for j in range(grid_num_particles[cell_to_check]):
p_j = grid2particles[cell_to_check, j]
if nb_i < max_num_neighbors and p_j != p_i and (
pos_i - positions[p_j]).norm() < neighbor_radius:
particle_neighbors[p_i, nb_i] = p_j
nb_i += 1
particle_num_neighbors[p_i] = nb_i
@ti.kernel
def substep():
# compute lambdas
# Eq (8) ~ (11)
for p_i in positions:
pos_i = positions[p_i]
grad_i = ti.Vector([0.0, 0.0, 0.0])
sum_gradient_sqr = 0.0
density_constraint = 0.0
for j in range(particle_num_neighbors[p_i]):
p_j = particle_neighbors[p_i, j]
if p_j < 0:
break
pos_ji = pos_i - positions[p_j]
grad_j = spiky_gradient(pos_ji, h)
grad_i += grad_j
sum_gradient_sqr += grad_j.dot(grad_j)
# Eq(2)
density_constraint += poly6_value(pos_ji.norm(), h)
# Eq(1)
density_constraint = (mass * density_constraint / rho0) - 1.0
sum_gradient_sqr += grad_i.dot(grad_i)
lambdas[p_i] = (-density_constraint) / (sum_gradient_sqr +
lambda_epsilon)
# compute position deltas
# Eq(12), (14)
for p_i in positions:
pos_i = positions[p_i]
lambda_i = lambdas[p_i]
pos_delta_i = ti.Vector([0.0, 0.0, 0.0])
for j in range(particle_num_neighbors[p_i]):
p_j = particle_neighbors[p_i, j]
if p_j < 0:
break
lambda_j = lambdas[p_j]
pos_ji = pos_i - positions[p_j]
scorr_ij = compute_scorr(pos_ji)
pos_delta_i += (lambda_i + lambda_j + scorr_ij) * \
spiky_gradient(pos_ji, h)
pos_delta_i /= rho0
position_deltas[p_i] = pos_delta_i
# apply position deltas
for i in positions:
positions[i] += position_deltas[i]
@ti.kernel
def epilogue():
# confine to boundary
for i in positions:
pos = positions[i]
positions[i] = confine_position_to_boundary(pos)
# update velocities
for i in positions:
velocities[i] = (positions[i] - old_positions[i]) / time_delta
# no vorticity/xsph because we cannot do cross product in 2D...
def run_pbf():
prologue()
for _ in range(pbf_num_iters):
substep()
epilogue()
@ti.kernel
def init():
init_pos = ti.Vector([10.0,10.0,10.0])
cube_size = 20
spacing = 1
num_per_row = (int) (cube_size // spacing) + 1
num_per_floor = num_per_row * num_per_row
for i in range(num_particles):
floor = i // (num_per_floor)
row = (i % num_per_floor) // num_per_row
col = (i % num_per_floor) % num_per_row
positions[i] = ti.Vector([col*spacing, floor*spacing, row*spacing]) + init_pos
def T(a):
if dim == 2:
return a
phi, theta = np.radians(28), np.radians(32)
a = a - 0.5
x, y, z = a[:, 0], a[:, 1], a[:, 2]
c, s = np.cos(phi), np.sin(phi)
C, S = np.cos(theta), np.sin(theta)
x, z = x * c + z * s, z * c - x * s
u, v = x, y * C + z * S
return np.array([u, v]).swapaxes(0, 1) + 25
def main():
init()
# init_particles()
print(f'boundary={boundary} grid={grid_size} cell_size={cell_size}')
gui = ti.GUI('PBF3D', screen_res, background_color = 0xffffff)
while gui.running and not gui.get_event(gui.ESCAPE):
run_pbf()
pos = positions.to_numpy()
# print(pos)
export_file = ""
if export_file:
writer = ti.tools.PLYWriter(num_vertices=num_particles)
writer.add_vertex_pos(pos[:, 0], pos[:, 1], pos[:, 2])
writer.export_frame(gui.frame, export_file)
gui.circles(T(pos)/100.0, radius=3, color=0x66ccff)
gui.show()
if __name__ == '__main__':
main()
边栏推荐
- PICT 生成正交测试用例教程
- mamp下缺少pcntl扩展的解决办法,Fatal error: Call to undefined function pcntl_signal()
- Redis introduction complete tutorial: detailed explanation of ordered collection
- [try to hack] wide byte injection
- 图片懒加载的原理
- Notepad++ -- editing skills
- Redis:Redis消息的发布与订阅(了解)
- Redis getting started complete tutorial: Key Management
- Unity Xiuxian mobile game | Lua dynamic sliding function (specific implementation of three source codes)
- Notepad++--编辑的技巧
猜你喜欢
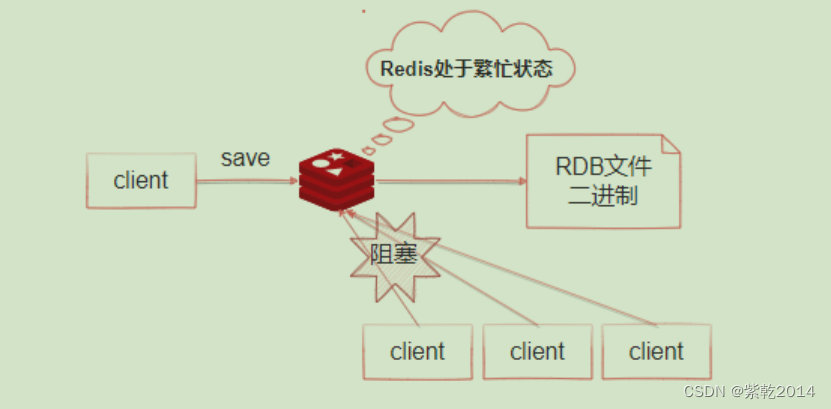
Persistence mechanism of redis
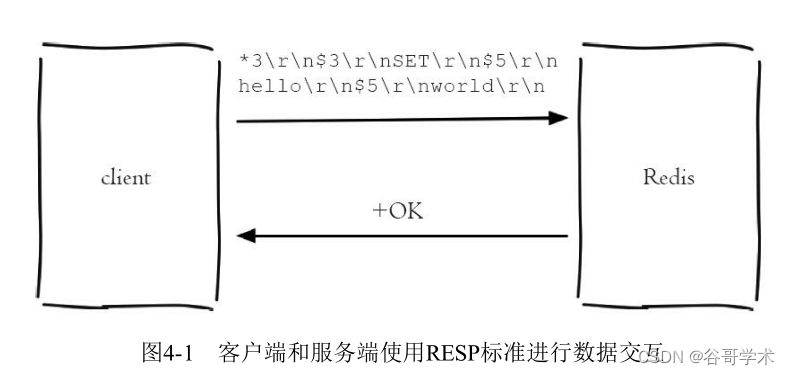
Redis introduction complete tutorial: client communication protocol
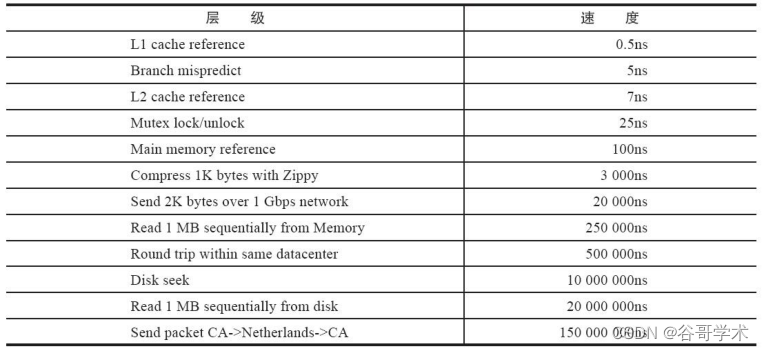
A complete tutorial for getting started with redis: getting to know redis for the first time
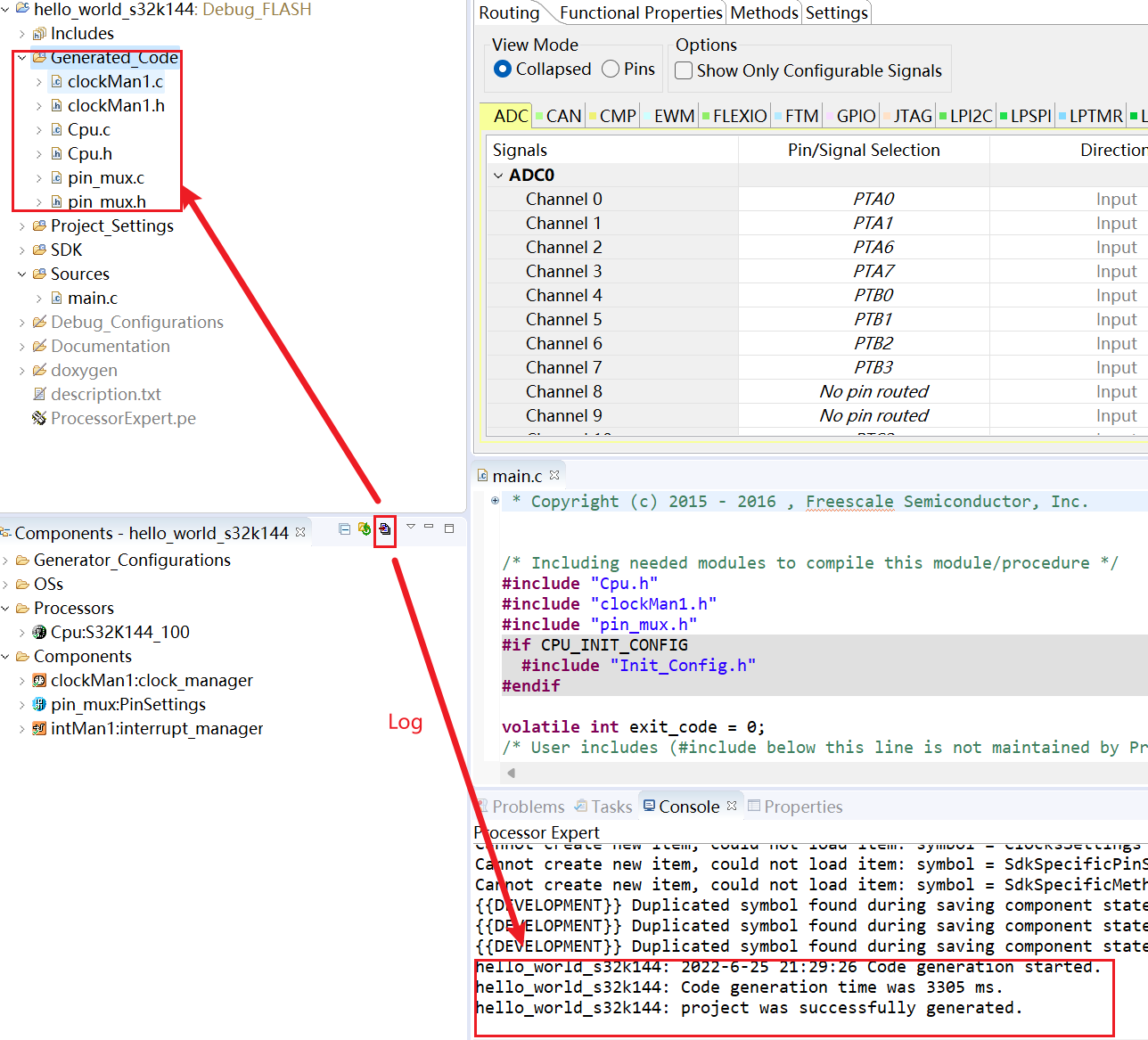
S32 Design Studio for ARM 2.2 快速入门

EditPlus--用法--快捷键/配置/背景色/字体大小
Redis入门完整教程:Pipeline
Analysis of the self increasing and self decreasing of C language function parameters
![[graph theory] topological sorting](/img/2c/238ab5813fb46a3f14d8de465b8999.png)
[graph theory] topological sorting
![[roommate learned to use Bi report data processing in the time of King glory in one game]](/img/06/22dde3fcc0456bd230e1d0cde339ec.png)
[roommate learned to use Bi report data processing in the time of King glory in one game]
Google Earth engine (GEE) - globfire daily fire data set based on mcd64a1
随机推荐
JS 3D explosive fragment image switching JS special effect
Notepad++--编辑的技巧
【ODX Studio编辑PDX】-0.2-如何对比Compare两个PDX/ODX文件
Analysis of environmental encryption technology
Servlet+JDBC+MySQL简单web练习
Advanced area of attack and defense world misc 3-11
Sword finger offer 65 Add without adding, subtracting, multiplying, dividing
时间 (计算)总工具类 例子: 今年开始时间和今年结束时间等
Redis入门完整教程:Redis Shell
Principle of lazy loading of pictures
A complete tutorial for getting started with redis: getting to know redis for the first time
Serial port data frame
Unity Xiuxian mobile game | Lua dynamic sliding function (specific implementation of three source codes)
Pagoda 7.9.2 pagoda control panel bypasses mobile phone binding authentication bypasses official authentication
A complete tutorial for getting started with redis: redis usage scenarios
Create Ca and issue certificate through go language
PICT 生成正交测试用例教程
A complete tutorial for getting started with redis: hyperloglog
[try to hack] wide byte injection
该如何去选择证券公司,手机上开户安不安全