当前位置:网站首页>[Practice 1] Diabetes Genetic Risk Detection Challenge [IFLYTEK Open Platform]
[Practice 1] Diabetes Genetic Risk Detection Challenge [IFLYTEK Open Platform]
2022-08-05 05:31:00 【Mosu playing computer】
文章目录
完成coggle任务
任务1:报名比赛
步骤1:报名比赛http://challenge.xfyun.cn/topic/info?type=diabetes&ch=ds22-dw-zmt05
赛事
在这次比赛中,您需要通过训练数据集构建糖尿病遗传风险预测模型,然后预测出测试数据集中个体是否患有糖尿病.对于测试数据集当中的个体,您必须预测其是否患有糖尿病(患有糖尿病:1,未患有糖尿病:0),预测值只能是整数1或者0.
训练集(比赛训练集.csv)一共有5070条数据,用于构建您的预测模型(您可能需要先进行数据分析).数据的字段有编号、性别、出生年份、体重指数、糖尿病家族史、舒张压、口服耐糖量测试、胰岛素释放实验、肱三头肌皮褶厚度、患有糖尿病标识(最后一列),您也可以通过特征工程技术构建新的特征.
测试集(比赛测试集.csv)一共有1000条数据,用于验证预测模型的性能.数据的字段有编号、性别、出生年份、体重指数、糖尿病家族史、舒张压、口服耐糖量测试、胰岛素释放实验、肱三头肌皮褶厚度.
提交说明
对于测试数据集当中的个体,您必须预测其是否患有糖尿病(患有糖尿病:1,未患有糖尿病:0),预测值只能是整数1或者0.提交的数据应该具有如下格式:
uuid,label
1,0
2,1
3,1
…
本次比赛中,预测模型的结果文件需要命名成:预测结果.csv,然后提交.请确保您提交的文件格式规范.
步骤2:下载比赛数据(点击比赛页面的赛题数据)
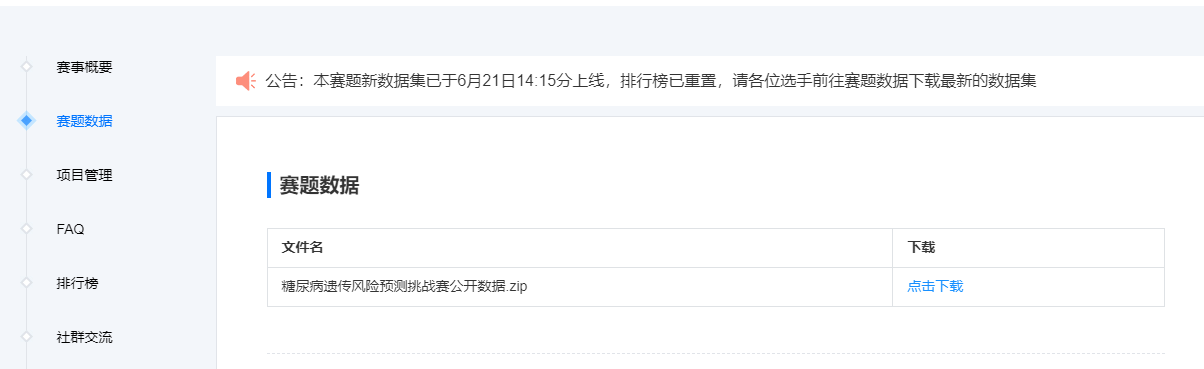
字段说明
编号:标识个体身份的数字;
性别:1表示男性,0表示女性;
出生年份:出生的年份;
体重指数:体重除以身高的平方,单位kg/m2;
糖尿病家族史:标识糖尿病的遗传特性,记录家族里面患有糖尿病的家属,分成三种标识,分别是父母有一方患有糖尿病、叔叔或者姑姑有一方患有糖尿病、无记录;
舒张压:心脏舒张时,动脉血管弹性回缩时,产生的压力称为舒张压,单位mmHg;
口服耐糖量测试:诊断糖尿病的一种实验室检查方法.比赛数据采用120分钟耐糖测试后的血糖值,单位mmol/L;
胰岛素释放实验:空腹时定量口服葡萄糖刺激胰岛β细胞释放胰岛素.比赛数据采用服糖后120分钟的血浆胰岛素水平,单位pmol/L;
肱三头肌皮褶厚度:在右上臂后面肩峰与鹰嘴连线的重点处,夹取与上肢长轴平行的皮褶,纵向测量,单位cm;
患有糖尿病标识:数据标签,1表示患有糖尿病,0表示未患有糖尿病.
步骤3:解压比赛数据,并使用pandas进行读取;
import pandas as pd
train_df = pd.read_csv("./比赛训练集.csv", encoding='gbk')
test_df = pd.read_csv("./比赛测试集.csv", encoding='gbk')
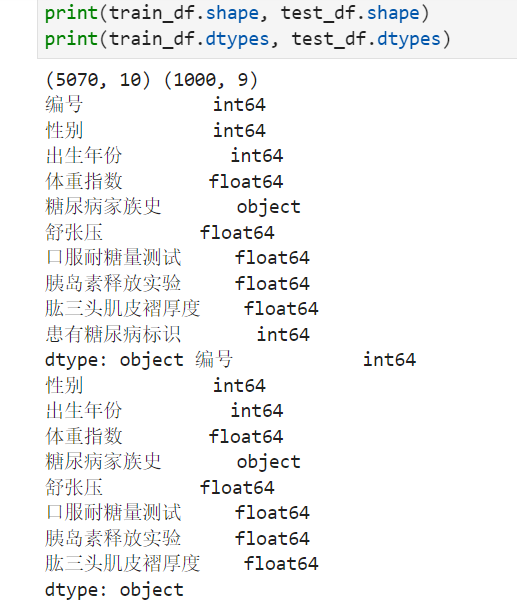
print(train_df.shape, test_df.shape)
print(train_df.dtypes, test_df.dtypes)
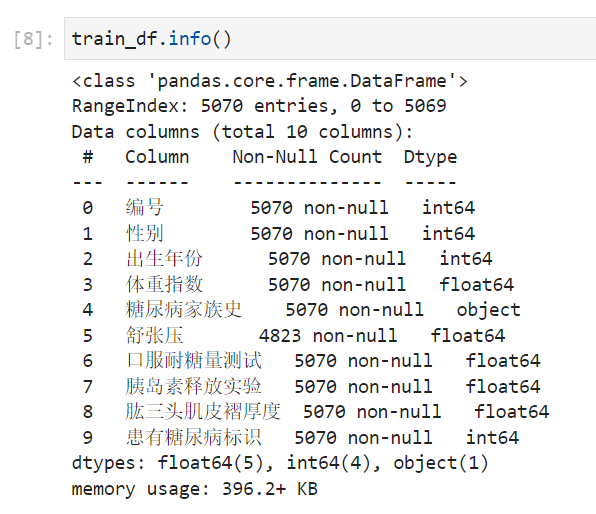
步骤4:查看训练集和测试集字段类型,并将数据读取代码写到博客;
任务2:比赛数据分析
步骤1:统计字段的缺失值,计算缺失比例;
通过缺失值统计,训练集和测试集的缺失值分布是否一致?
通过缺失值统计,有没有缺失比例很高的列?
#缺失值计算
train_df.isnull().mean(0)
test_df.isnull().mean(0)
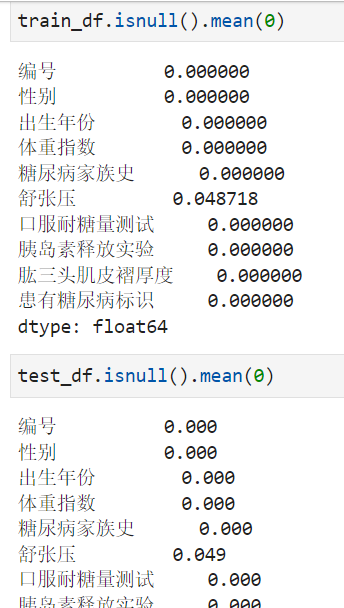
It can be seen that the field of diastolic blood pressure is missing 4.9%.The test set and training set have similar proportions.
步骤2:分析字段的类型;
从info和head(查看前5行的数据)可以看出,体重,舒张压,口服耐糖量测试 ,胰岛素释放实验,Triceps skinfold thickness is continuous,The rest are integers(Diabetes sign is0 1).The family history column is a note(In the later stage, this feature should be processed by the character type and can be processed as an integer type to join the use of the model.).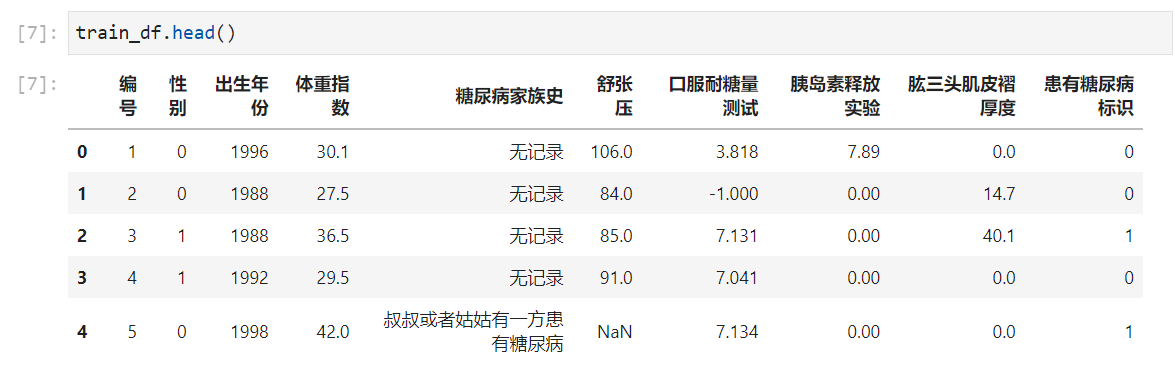
步骤3:计算字段相关性;
通过.corr()计算字段之间的相关性;
# 相关性计算
train_df.corr()
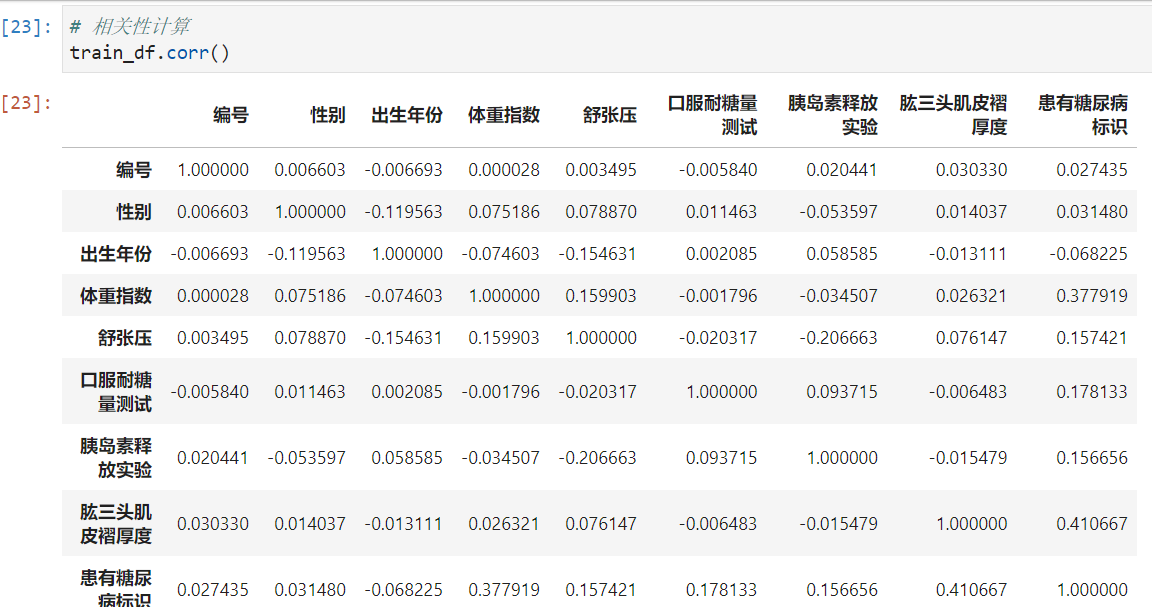
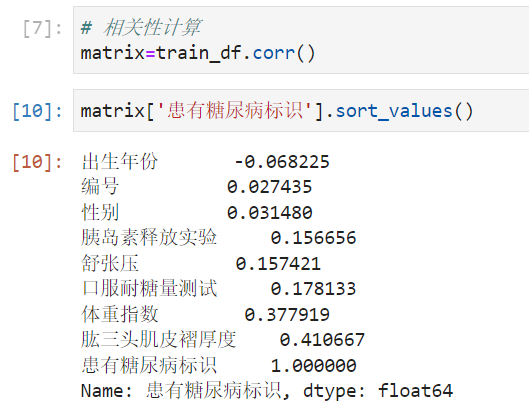
Which fields are most relevant to labels
- 可以看出 Triceps skinfold thickness and body mass coefficient are more closely related to diabetes.
可视化
尝试使用其他可视化方法将字段 与 标签的分布差异进行可视化;
The following is also a chart to look at the distribution of diabetes.
之前没用过seaborn,boxplotShould be a box plot,countplot,和violinplot就不知道了
查了一下,countplot就是直方图,violinplot是violin-style boxplot.
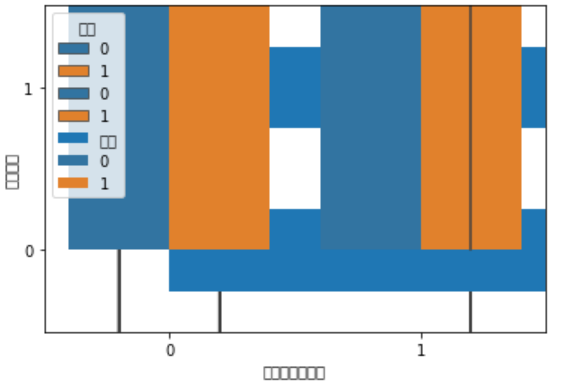
The reason for the above picture is1.seabornSeveral pictures are stacked together,2.I didn't switch the language setting to Chinese and it didn't work(之后加入设置代码,In this way, the symbols of Chinese and negative axes can be displayed normally),Picture set stolen from teachersvg语句,更清楚一些
import seaborn as sns
import matplotlib.pyplot as plt
plt.rcParams['font.family'] = ['SimHei'] #用来正常显示中文标签
plt.rcParams['axes.unicode_minus'] = False #用来正常显示负号
from IPython.display import set_matplotlib_formats
set_matplotlib_formats('svg')#Display image format
# train_df['性别'].value_counts().plot(kind='barh')# barh水平的,kind类型
# sns.countplot(x='患有糖尿病标识', hue='性别', data=train_df)#hue分列 value_counts和 countplot其实是一样的
# sns.boxplot(y='出生年份', x='患有糖尿病标识', hue='性别', data=train_df)
sns.violinplot(y="体重指数", x="患有糖尿病标识", hue="性别", data=train_df)
Annotate image statements in turn,得到下列图片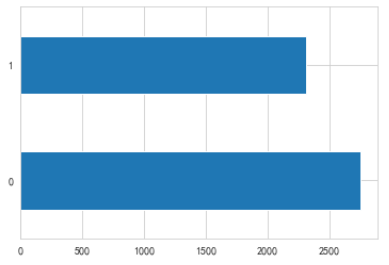
1是男性,0是女性.more women
barh 水平的
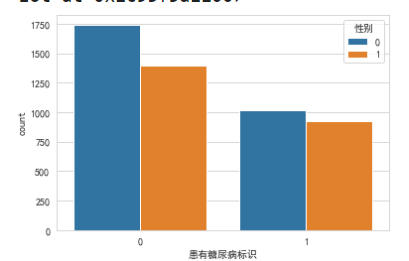
横坐标轴0don't have diabetes,blue vertical bar0是女性.Anyway, more women,This also does not see a relationship between diabetes and gender.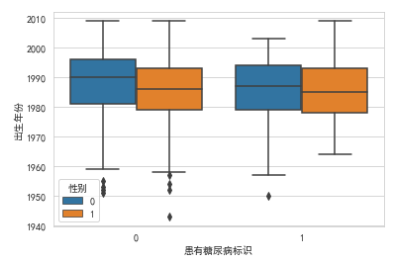
Men's yellow bars is normal distribution,The female blue bar has moved up a bit,not very normal.man without diabetes,There are a few very long-lived points(离群点).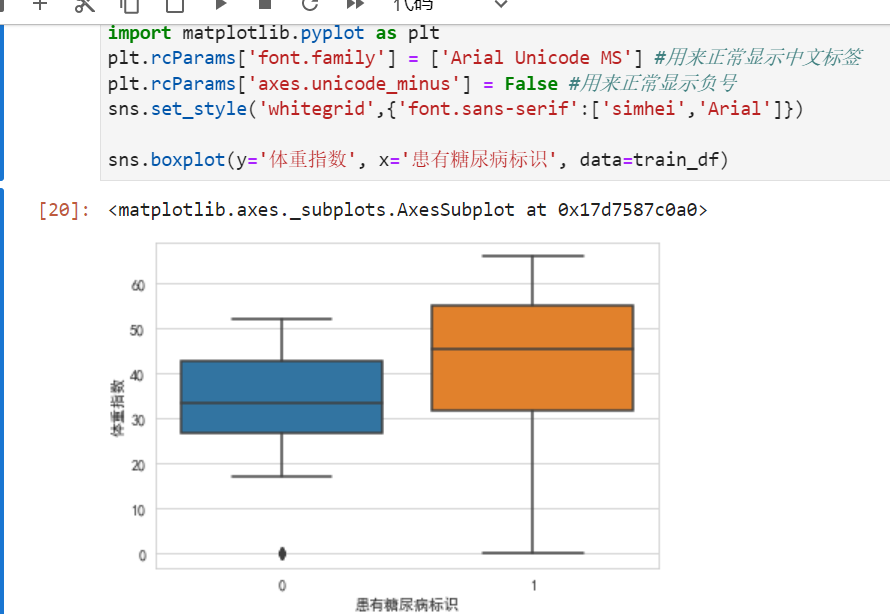
People with diabetes have a slightly higher weight distribution than normal.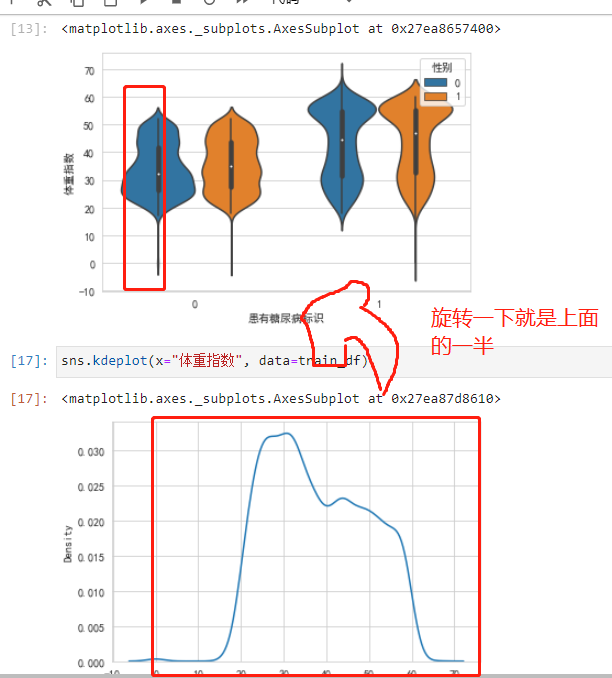
The violin plot is actually the measured density histogram,对称的,Just look at half.
scatterplot 散点图,xy都是数值型
然后 Each figure has some 属性,like what paddingfill 还有 alpha透明度,You can go to the official website to learn.
tips:根据xyThe data type of the axis depends on which graph to use
Based on these, we can judge whether 做交叉特征(Addition, subtraction, multiplication and division of two columns),can be separated
补充学习 b站7.1录播
groupby
apply
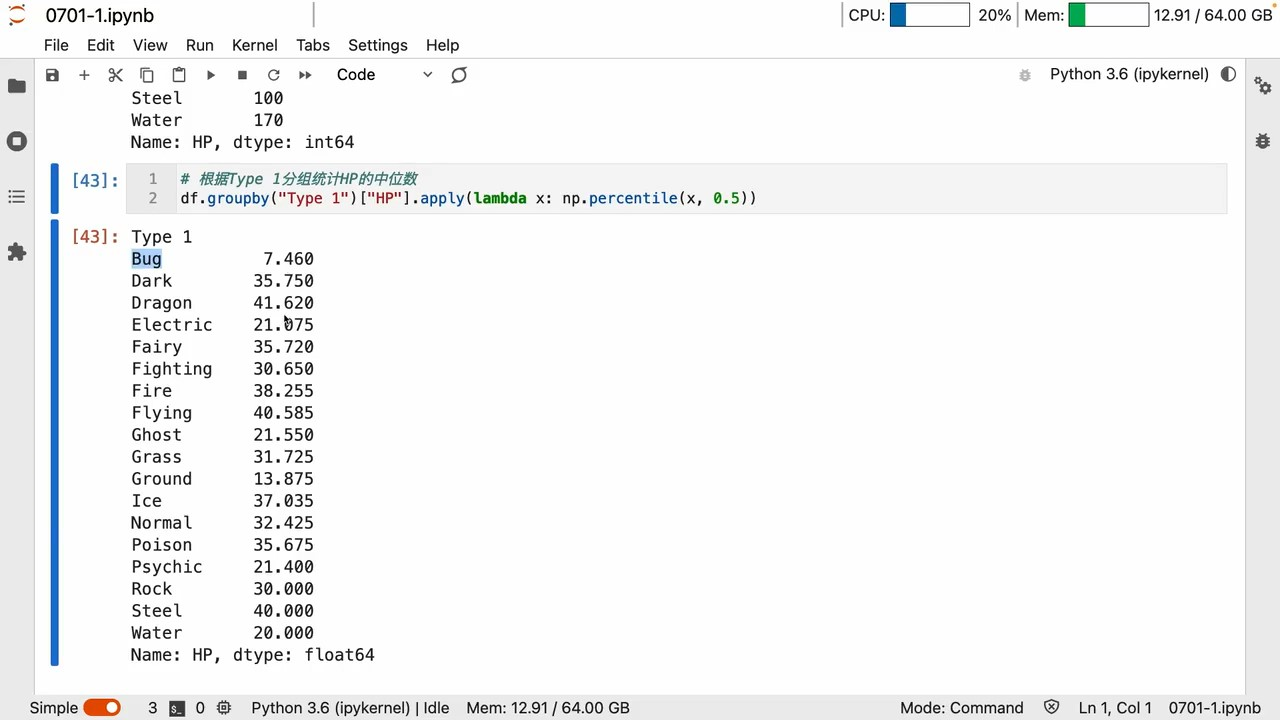
apply用 lambda Can be customized a lot
agg
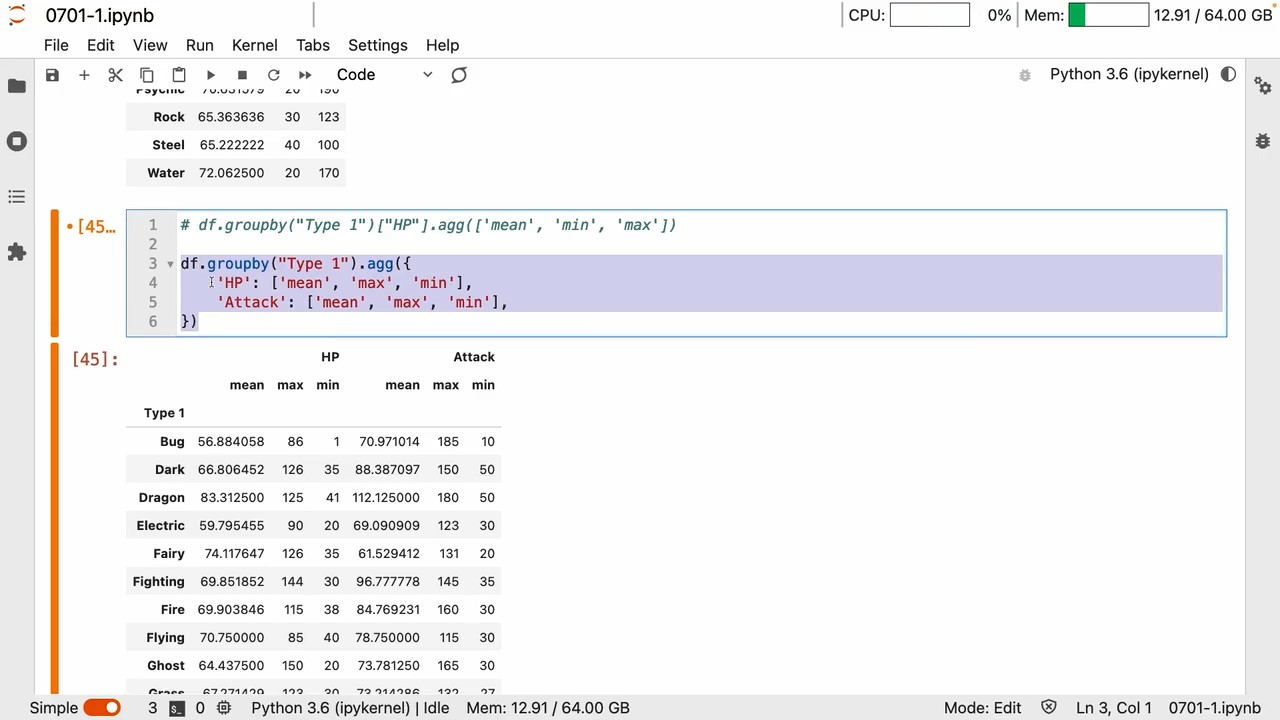
- groupby 和 agg Can achieve elegant aggregation effects
map、transform
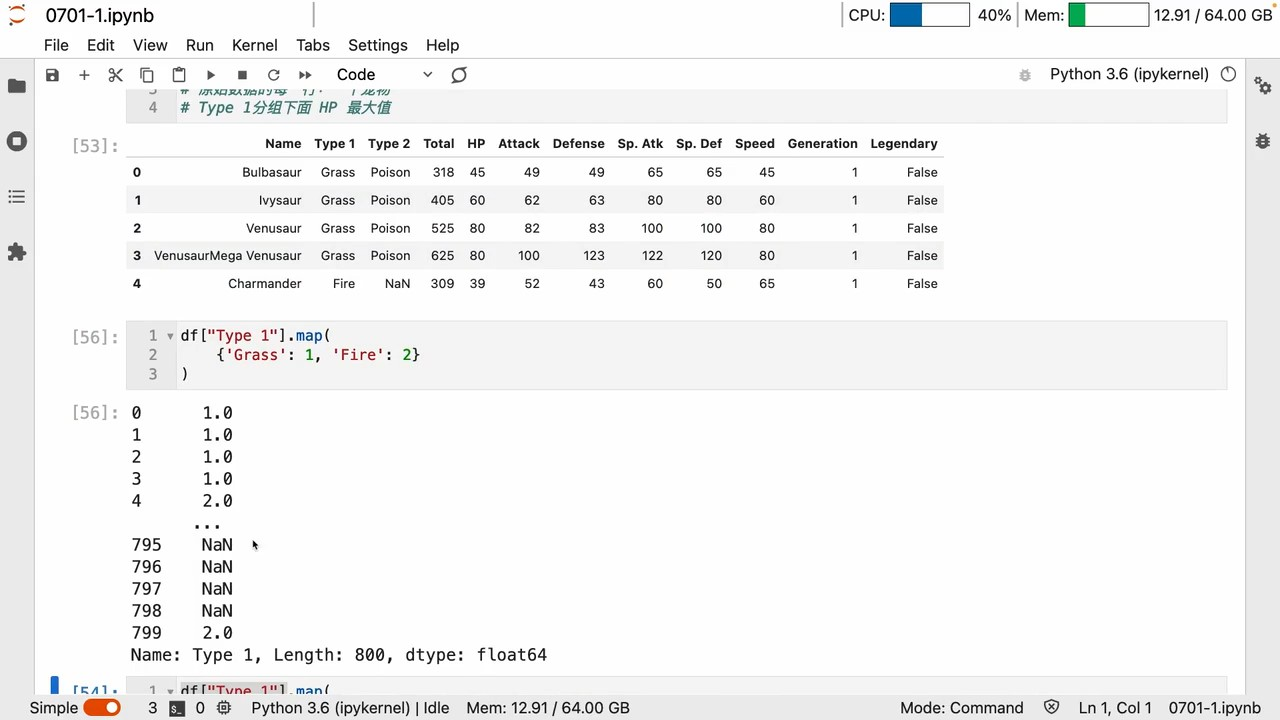 mapCan map changes 键值
mapCan map changes 键值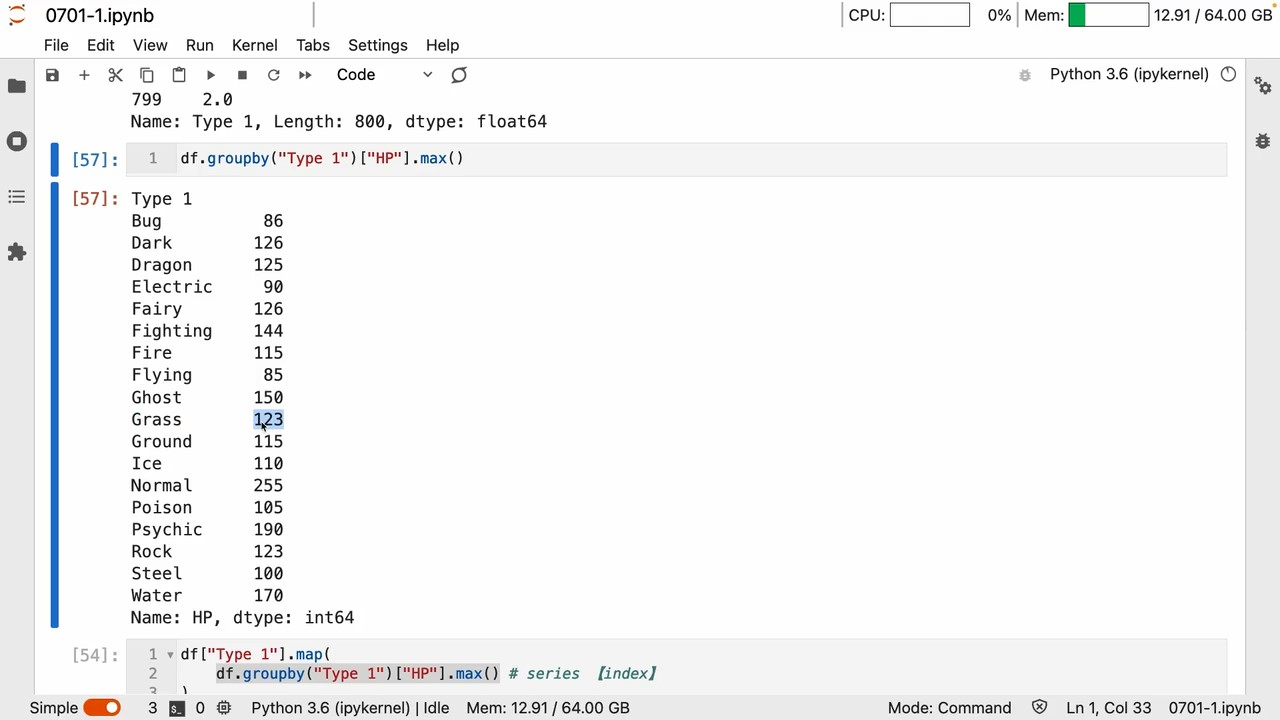
groupby A series of operations is to get the key-value relationship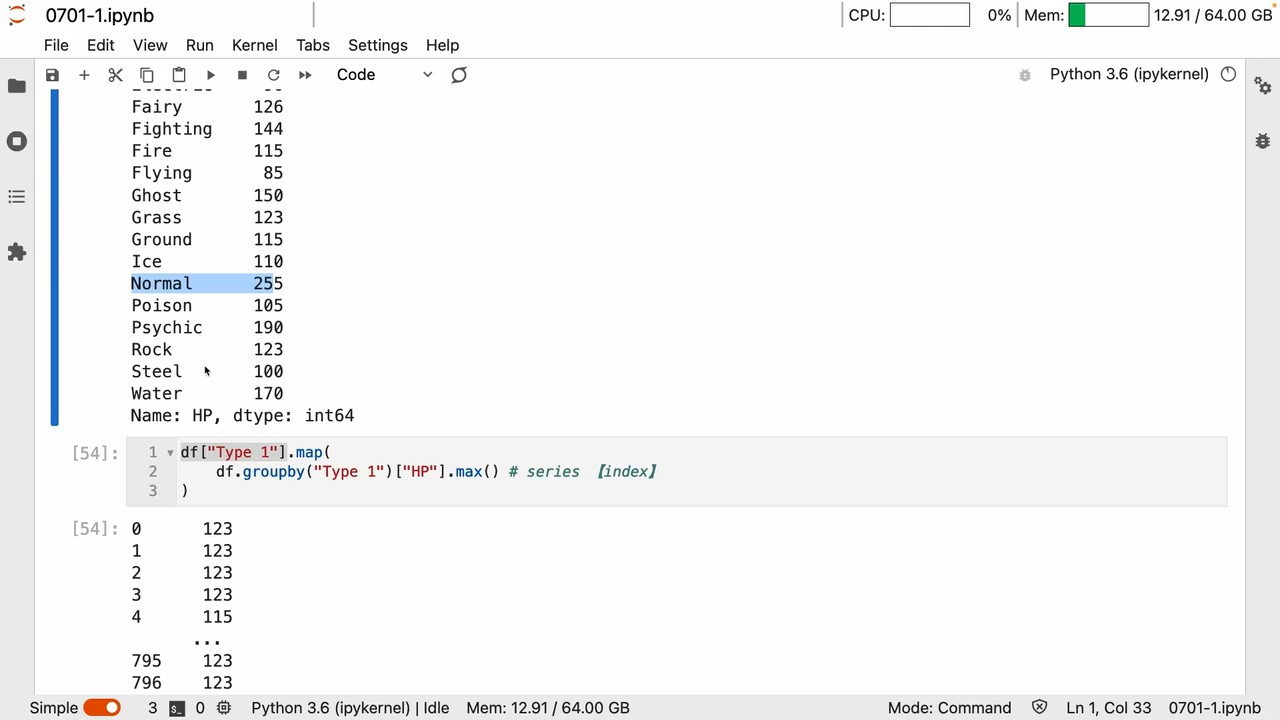
所以 map里面写个groupby Naturally, you can map and replace
实例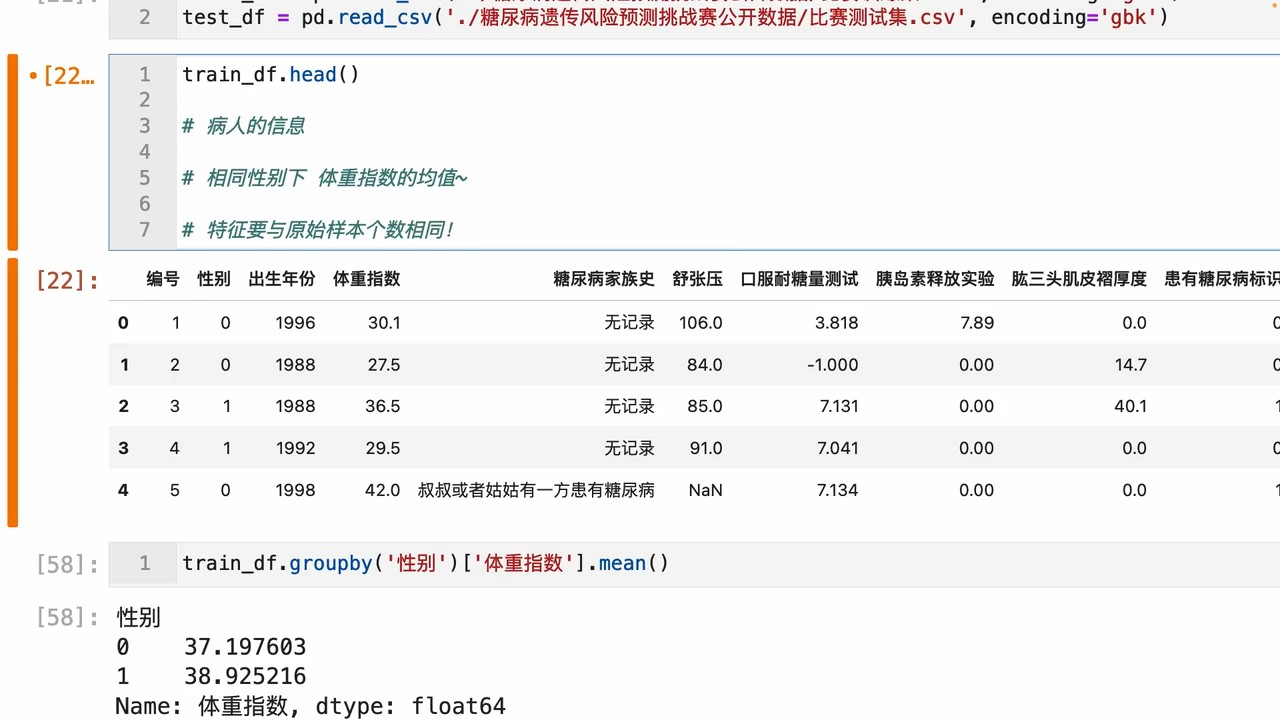
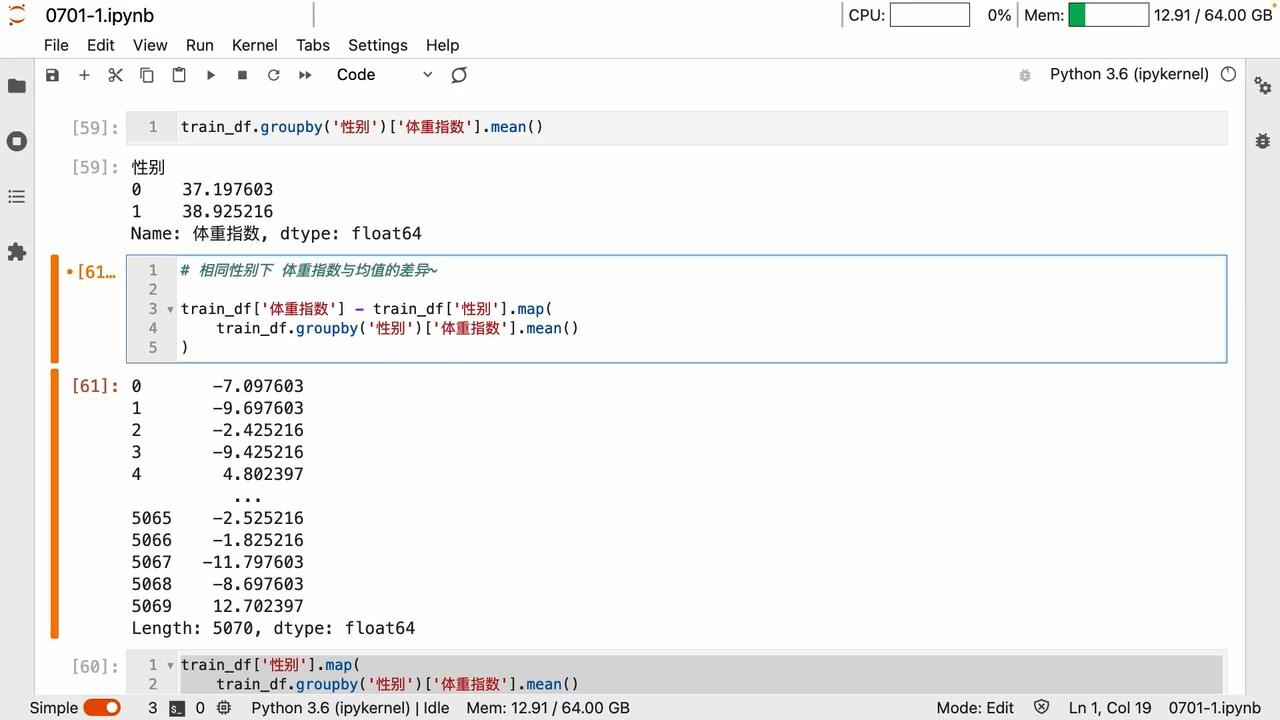
A list of new features with suitable dimensions can be generated in this way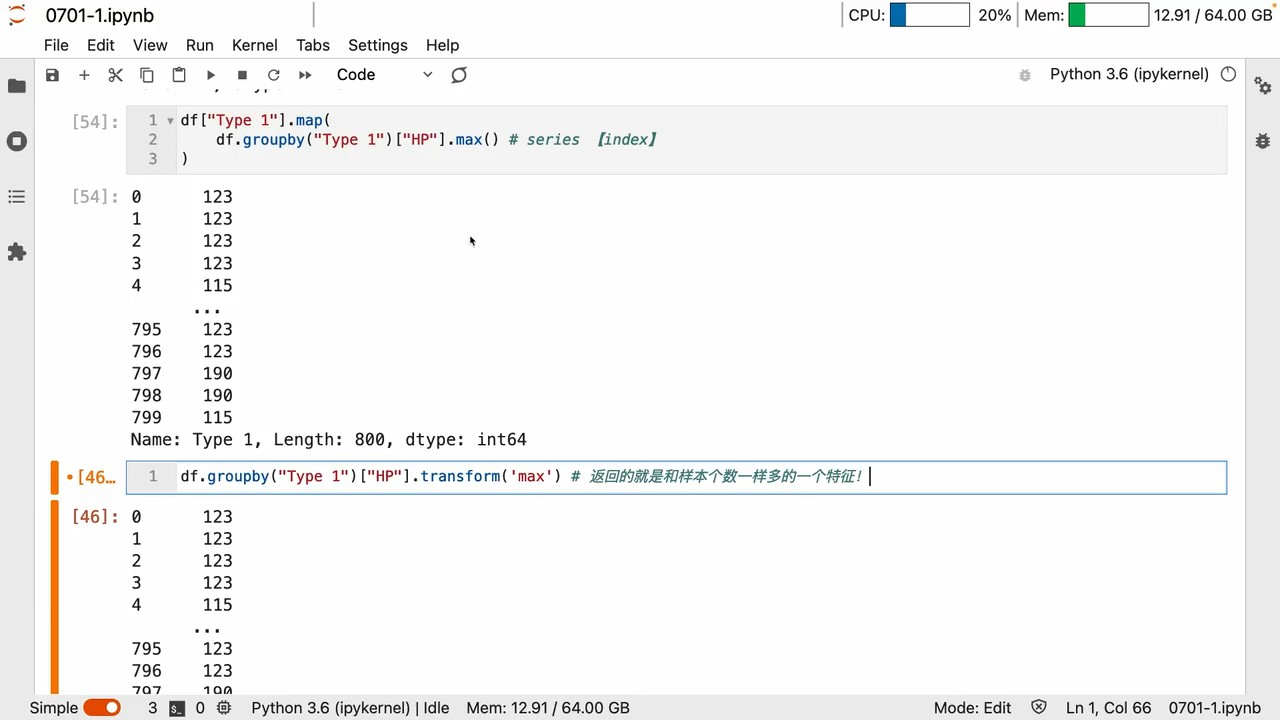
可视化
See step 3 above
特征工程
onehot

labelEncoder
The usual categorical variables arelabelEncoder即可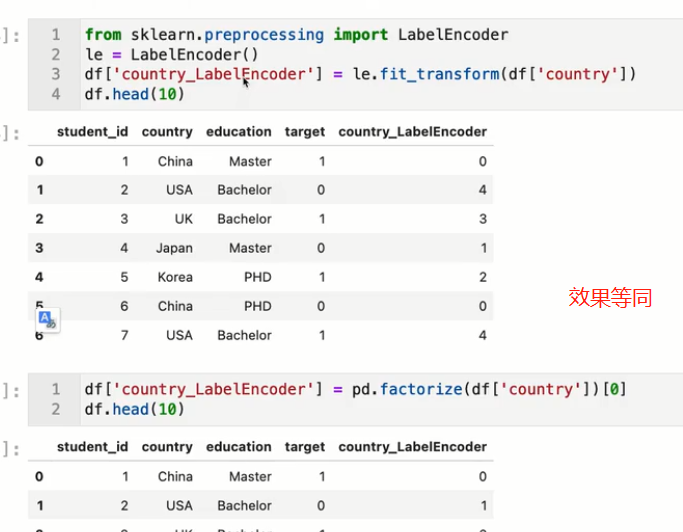
有序变量
need to preserve order 有序变量 用map
任务3:逻辑回归尝试
步骤1:导入sklearn中的逻辑回归;
import pandas as pd
from sklearn.linear_model import LogisticRegression
from sklearn.preprocessing import MinMaxScaler
from sklearn.pipeline import make_pipeline
导入对应的包
步骤1.5 数据预处理
I think because family history can be regarded as an ordinal variable according to common sense,而且‘叔叔或者姑姑有一方患有糖尿病’和‘叔叔或姑姑有一方患有糖尿病’其实(含义一样)是一类,如果直接用labelEncoderwill become two types,是不对的,所以用map处理.
train_df = pd.read_csv('./糖尿病遗传风险预测挑战赛公开数据/比赛训练集.csv', encoding='gbk')
test_df = pd.read_csv('./糖尿病遗传风险预测挑战赛公开数据/比赛测试集.csv', encoding='gbk')
dict_糖尿病家族史 = {
'无记录': 0,
'叔叔或姑姑有一方患有糖尿病': 1,
'叔叔或者姑姑有一方患有糖尿病': 1,
'父母有一方患有糖尿病': 2
}
train_df['糖尿病家族史'] = train_df['糖尿病家族史'].map(dict_糖尿病家族史)
test_df['糖尿病家族史'] = test_df['糖尿病家族史'].map(dict_糖尿病家族史)
train_df['舒张压'].fillna(89, inplace=True)
test_df['舒张压'].fillna(89, inplace=True)
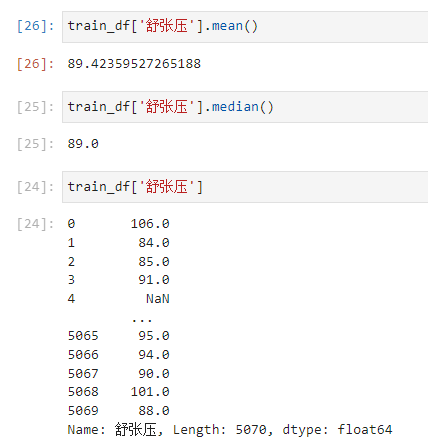
Diastolic blood pressure is either the mean or the median89this value here,And observe that the original data are integers,所以就用89了.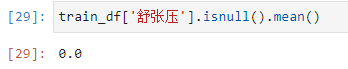
补全之后,没有缺失值了.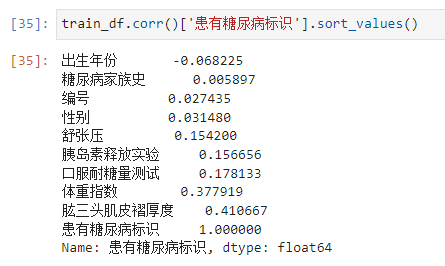
Correlation between family history and diabetes markers,这么低吗.(意外,Mingming EarthOL里面,Genetics is an important factor in causing diabetes)
步骤2:使用训练集和逻辑回归进行训练,并在测试集上进行预测;
读数据-》Convert an ordinal variable to a numeric variable(利用map)
Based on previous data analysis,(4.9%absence does not remove),So fill in missing values
from sklearn.linear_model import LogisticRegression as LR
from sklearn.model_selection import cross_val_score
from sklearn.feature_selection import SelectFromModel
import numpy as np
import pandas as pd
from sklearn.metrics import accuracy_score
from sklearn.preprocessing import MinMaxScaler
from sklearn.pipeline import make_pipeline
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import cross_val_score
features=[f for f in train_df.columns if f not in ['编号','患有糖尿病标识','出生年份']]
X=train_df[features]
y=train_df['患有糖尿病标识']
Xtrain, Xtest, Ytrain, Ytest = train_test_split(X,y,test_size=0.2,random_state=2022)
#修正索引
for i in [Xtrain, Xtest, Ytrain, Ytest]:
i.index=range(i.shape[0])
#Confirm with images
l1 = []
l2 = []
l1test = []
l2test = []
for i in np.linspace(0.05,1,19):
lrl1 = LR(penalty="l1",solver="liblinear",C=i,max_iter=1000)
lrl2 = LR(penalty="l2",solver="liblinear",C=i,max_iter=1000)
lrl1 = lrl1.fit(Xtrain,Ytrain)
l1.append(accuracy_score(lrl1.predict(Xtrain),Ytrain))
l1test.append(accuracy_score(lrl1.predict(Xtest),Ytest))
lrl2 = lrl2.fit(Xtrain,Ytrain)
l2.append(accuracy_score(lrl2.predict(Xtrain),Ytrain))
l2test.append(accuracy_score(lrl2.predict(Xtest),Ytest))
graph = [l1,l2,l1test,l2test]
color = ["green","black","lightgreen","gray"]
label = ["L1","L2","L1test","L2test"]
plt.figure(figsize=(6,6))
for i in range(len(graph)):
plt.plot(np.linspace(0.05,1,19),graph[i],color[i],label=label[i])
plt.legend(loc=4)
plt.show()
步骤3:将步骤2预测的结果文件提交到比赛,截图分数;
我是看 l2正则化的逻辑回归,Then the performance on the test set is better,Then by reducing the space,found the bestC值,结果交上去,才71.(maybe i put 出生年份 reason for removal)It is also possible to identify the cause 谁知道呢
lrl2 = LR(penalty="l2",solver="liblinear",C=0.395,max_iter=1000)
lrl2 = lrl2.fit(Xtrain,Ytrain)
test_df['label'] = lrl2.predict(test_df.drop(['编号','出生年份'], axis=1))
test_df.rename({
'编号': 'uuid'}, axis=1)[['uuid', 'label']].to_csv('submit.csv', index=None)
步骤4:将训练集20%划分为验证集,在训练部分进行训练,在测试部分进行预测,调节逻辑回归的超参数;
(看了一下别人博客,nor stipulatedl1,l2,就默认l2,然后C=1e2,Then the rating is73)
步骤4:如果精度有提高,则重复步骤2和步骤3;如果没有提高,可以尝试树模型,重复步骤2、3;
#网格搜索
parameters = {
'splitter':('best','random')
,'criterion':("gini","entropy")
,"max_depth":[*range(1,10)]
# 'min_samples_leaf':[*range(1,50,5)]
# ,'min_impurity_decrease':[*np.linspace(0,0.5,20)]
}
clf = DecisionTreeClassifier(random_state=25)
GS = GridSearchCV(clf, parameters, cv=10)
GS.fit(Xtrain,Ytrain)
GS.best_params_
# GS.best_score_
#draw a curve
tr=[]
te=[]
for i in range(40):
clf=DecisionTreeClassifier(random_state=25
,max_depth=7
,criterion="entropy"
,min_samples_leaf=i+1
)
clf=clf.fit(Xtrain,Ytrain)
score_tr=clf.score(Xtrain,Ytrain)
score_te=cross_val_score(clf,X,y,cv=10).mean()
tr.append(score_tr)
te.append(score_te)
print(max(te),te.index(max(te)))
plt.plot(range(1,41),tr,color='red',label='train')
plt.plot(range(1,41),te,color='green',label='test')
plt.xticks(range(1,41))
plt.legend()
plt.show()
提交答案
clf = DecisionTreeClassifier(random_state=25
,min_samples_leaf=25
,max_depth=7
,criterion="entropy"
)
clf.fit(Xtrain,Ytrain)
score=cross_val_score(clf,X,y,cv=10).mean()
score
任务4:特征工程(使用pandas完成)
步骤1:统计每个性别对应的【体重指数】、【舒张压】平均值
train_df.groupby("性别").agg({
'体重指数':['mean'],'舒张压':['mean']})
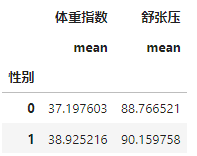
步骤2:计算每个患者与每个性别平均值的差异;
train_df['体重指数mean']=train_df['体重指数']-train_df['性别'].map(
train_df.groupby("性别")['体重指数'].mean()
)
train_df['舒张压mean']=train_df['舒张压']-train_df['性别'].map(
train_df.groupby("性别")['舒张压'].mean()
)
In terms of correlation,差不多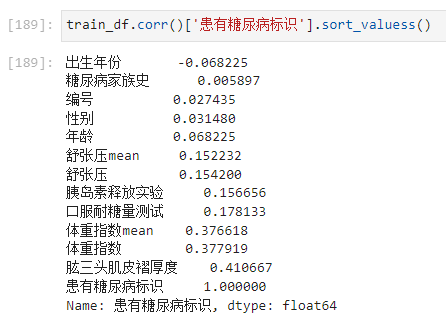
步骤3:在上述基础上将训练集20%划分为验证集,使用逻辑回归完成训练,精度是否有提高?
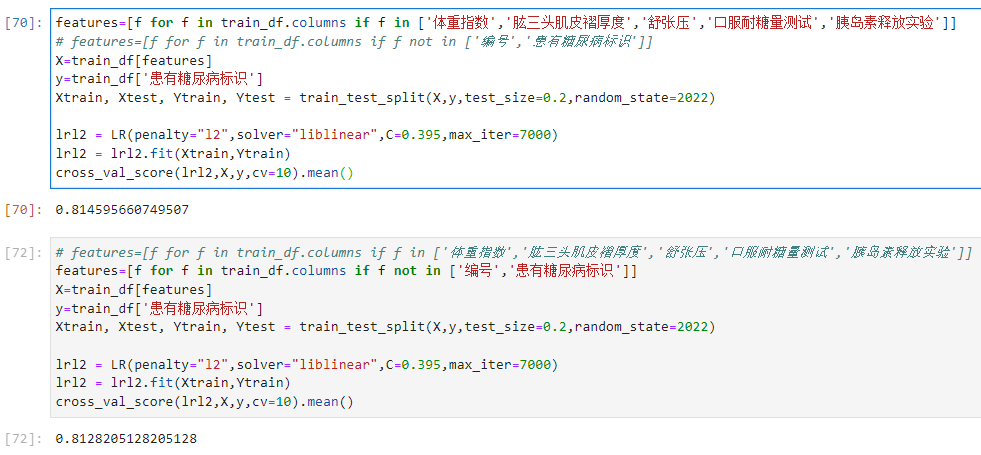
步骤4:思考字段含义,尝试新的特征,将你的尝试写入博客;
import pandas as pd
data={
'featureName':features,'importances':clf.feature_importances_.tolist()}
df = pd.DataFrame(data)
df=df.sort_values(by=['importances'],ascending=False)
df
新特征 好像没啥用,Decision trees look down on them.
任务5:特征筛选
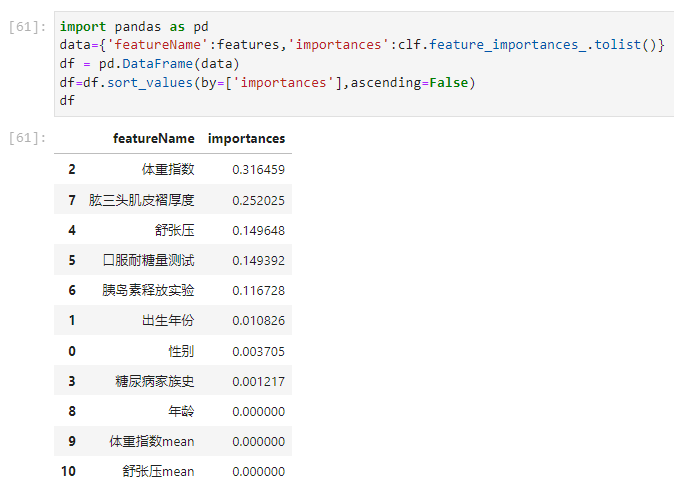
步骤1:使用树模型完成模型的训练,通过特征重要性筛选出Top5的特征;
From the picture above it can be seen that
体重指数 0.316459
肱三头肌皮褶厚度 0.252025
舒张压 0.149648
口服耐糖量测试 0.149392
胰岛素释放实验 0.116728
步骤2:使用筛选出的特征和逻辑回归进行训练,在验证集精度是否有提高?
提高了
步骤3:如果有提高,为什么?如果没有提高,为什么?

划重点了,Exam a little better.
But this is certainly not a significant improvement,The model is at its limit,That's it for logistic regression.
步骤4:将你的尝试写入博客;
任务6:高阶树模型
import pandas as pd
import numpy as np
import seaborn as sns
from sklearn.model_selection import KFold
import lightgbm as lgb
# 读取数据
train_df = pd.read_csv('./糖尿病遗传风险预测挑战赛公开数据/比赛训练集.csv', encoding='gbk')
test_df = pd.read_csv('./糖尿病遗传风险预测挑战赛公开数据/比赛测试集.csv', encoding='gbk')
# 基础特征工程
train_df['体重指数_round'] = train_df['体重指数'] // 10
test_df['体重指数_round'] = train_df['体重指数'] // 10
# //地板除 Python中两个斜杠即双斜杠(//)表示地板除,即先做除法(/),然后向下取整(floor).
train_df['口服耐糖量测试'] = train_df['口服耐糖量测试'].replace(-1, np.nan)
test_df['口服耐糖量测试'] = test_df['口服耐糖量测试'].replace(-1, np.nan)
# This feature has-1 懵逼了
dict_糖尿病家族史 = {
'无记录': 0,
'叔叔或姑姑有一方患有糖尿病': 1,
'叔叔或者姑姑有一方患有糖尿病': 1,
'父母有一方患有糖尿病': 2
}
train_df['糖尿病家族史'] = train_df['糖尿病家族史'].map(dict_糖尿病家族史)
test_df['糖尿病家族史'] = test_df['糖尿病家族史'].map(dict_糖尿病家族史)
train_df['糖尿病家族史'] = train_df['糖尿病家族史'].astype('category')
test_df['糖尿病家族史'] = train_df['糖尿病家族史'].astype('category')
train_df['性别'] = train_df['性别'].astype('category')
test_df['性别'] = train_df['性别'].astype('category')
# Does this conversion to a category have any special meaning?
train_df['年龄'] = 2022 - train_df['出生年份']
test_df['年龄'] = 2022 - test_df['出生年份']
#worked before,but it's no use
train_df['口服耐糖量测试_diff'] = train_df['口服耐糖量测试'] - train_df.groupby('糖尿病家族史').transform('mean')['口服耐糖量测试']
test_df['口服耐糖量测试_diff'] = test_df['口服耐糖量测试'] - test_df.groupby('糖尿病家族史').transform('mean')['口服耐糖量测试']
#It turns out that this feature needs to operate,rather than body weight and diastolic blood pressure
步骤1:安装LightGBM,并学习基础的使用方法;
安装一下
学一下
a translated document
官方文档
知乎博文
步骤2:将训练集20%划分为验证集,使用LightGBM完成训练,精度是否有提高?
用coogleRun the template code given by the teacher
# 模型交叉验证
def run_model_cv(model, kf, X_tr, y, X_te, cate_col=None):
train_pred = np.zeros( (len(X_tr), len(np.unique(y))) )
test_pred = np.zeros( (len(X_te), len(np.unique(y))) )
cv_clf = []
for tr_idx, val_idx in kf.split(X_tr, y):
x_tr = X_tr.iloc[tr_idx]; y_tr = y.iloc[tr_idx]
x_val = X_tr.iloc[val_idx]; y_val = y.iloc[val_idx]
call_back = [
lgb.early_stopping(50),
]
eval_set = [(x_val, y_val)]
model.fit(x_tr, y_tr, eval_set=eval_set, callbacks=call_back, verbose=-1)
cv_clf.append(model)
train_pred[val_idx] = model.predict_proba(x_val)
test_pred += model.predict_proba(X_te)
test_pred /= kf.n_splits
return train_pred, test_pred, cv_clf
```python
clf = lgb.LGBMClassifier(
max_depth=3,
n_estimators=4000,
n_jobs=-1,
verbose=-1,
verbosity=-1,
learning_rate=0.1,
)
train_pred, test_pred, cv_clf = run_model_cv(
clf, KFold(n_splits=5),
train_df.drop(['编号', '患有糖尿病标识'], axis=1),
train_df['患有糖尿病标识'],
test_df.drop(['编号'], axis=1),
)
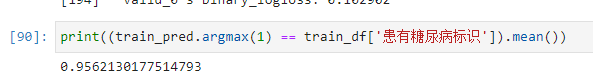
print((train_pred.argmax(1) == train_df['患有糖尿病标识']).mean())
test_df['label'] = test_pred.argmax(1)
test_df.rename({
'编号': 'uuid'}, axis=1)[['uuid', 'label']].to_csv('submit.csv', index=None)
步骤3:将步骤2预测的结果文件提交到比赛,截图分数;
步骤4:尝试调节搜索LightGBM的参数;
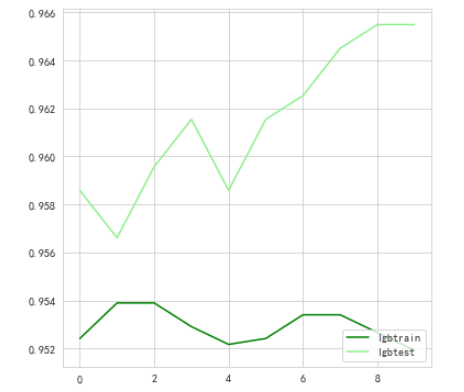
Looked at the effect of parameters of different depths
lgbtrain = []
lgbtest = []
for i in range(10):
clf = lgb.LGBMClassifier(
max_depth=i+1,
n_estimators=4000,
n_jobs=-1,
verbose=-1,
verbosity=-1,
learning_rate=0.1,
)
train_pred, test_pred, cv_clf = run_model_cv(
clf, KFold(n_splits=5),
Xtrain,
Ytrain,
Xtest,
)
lgbtrain.append((train_pred.argmax(1) == Ytrain).mean())
lgbtest.append((test_pred.argmax(1) == Ytest).mean())
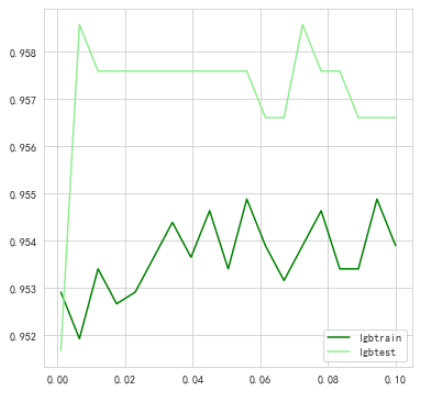
Look at the effect of parameters with different learning rates
lgbtrain = []
lgbtest = []
# for i in np.linspace(0.39,0.40,19):
for i in np.linspace(0.001,0.1,19):
clf = lgb.LGBMClassifier(
max_depth=2,
n_estimators=4000,
n_jobs=-1,
verbose=-1,
verbosity=-1,
learning_rate=i,
)
train_pred, test_pred, cv_clf = run_model_cv(
clf, KFold(n_splits=5),
Xtrain,
Ytrain,
Xtest,
)
lgbtrain.append((train_pred.argmax(1) == Ytrain).mean())
lgbtest.append((test_pred.argmax(1) == Ytest).mean())
graph = [lgbtrain,lgbtest]
color = ["green","lightgreen"]
label = ["lgbtrain","lgbtest"]
plt.figure(figsize=(6,6))
for i in range(len(graph)):
plt.plot(np.linspace(0.001,0.1,19),graph[i],color[i],label=label[i])
plt.legend(loc=4)
plt.show()
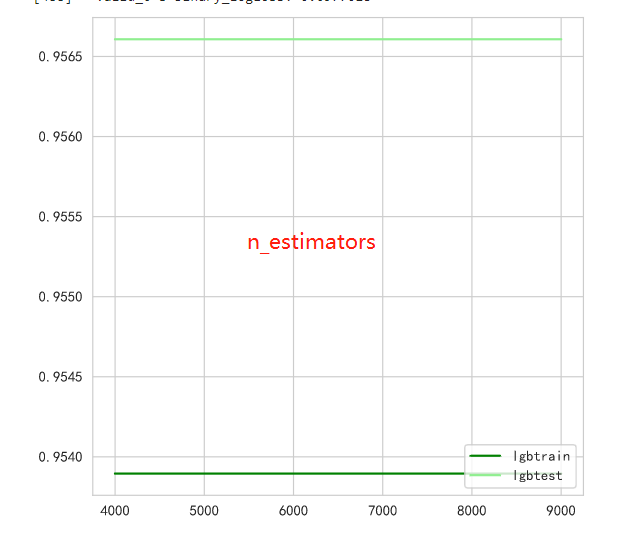
步骤5:将步骤4调参之后的模型从新训练,将最新预测的结果文件提交到比赛,截图分数;
蚌埠住了,更低了
任务7:多折训练与集成
可以参考 大佬的https://blog.csdn.net/qq_39473431/article/details/125478123
And the big guy mentioned https://www.kaggle.com/code/arthurtok/introduction-to-ensembling-stacking-in-python/notebook
步骤1:使用KFold完成数据划分;
KFoldThe principle of dividing the data set:根据n_splitDivide directly.already divided
步骤2:使用StratifiedKFold完成数据划分;
StratifiedKFoldThe principle of dividing the data set:The divided training set and validation set类别The distribution is as close as possible to the original dataset
使用
Practice in data races
理论
调参
步骤3:使用StratifiedKFold配合LightGBM完成模型的训练和预测
coggleThe example given isKFold的,修改相应代码即可
步骤4:在步骤3训练得到了多少个模型,对测试集多次预测,将最新预测的结果文件提交到比赛,截图分数;
步骤4:使用交叉验证训练5个机器学习模型(svm、lr等),使用stacking完成集成,将最新预测的结果文件提交到比赛,截图分数;
集成
make a good model,Have good data first,The quality of the data determines the upper limit of your model's performance;其次,要做好特征工程,Under conditions that cannot alter data quality,特征工程是重中之重;最后,建立模型.
There are three mainstream model ideas,一种是Bagging,The representative model is random forest;一种是Boosting,代表模型是GBDT、Xgboost、lightGBM;还一种是stacking或blending.前2models can be adjusted.
总结
可以像这样
Describe the difficulties and the process
边栏推荐
- spark-DataFrame数据插入mysql性能优化
- [Go through 10] sklearn usage record
- Mysql-连接https域名的Mysql数据源踩的坑
- 学习总结week2_5
- 学习总结day5
- ES6 新特性:Class 的继承
- 如何编写一个优雅的Shell脚本(一)
- 【零基础开发NFT智能合约】如何使用工具自动生成NFT智能合约带白名单可Mint无需写代码
- In Opencv, imag=cv2.cvtColor(imag,cv2.COLOR_BGR2GRAY) error: error:!_src.empty() in function 'cv::cvtColor'
- [Go through 8] Fully Connected Neural Network Video Notes
猜你喜欢

pycharm中调用Matlab配置:No module named ‘matlab.engine‘; ‘matlab‘ is not a package
IDEA 配置连接数据库报错 Server returns invalid timezone. Need to set ‘serverTimezone‘ property.
Lecture 4 Backpropagation Essays
软件设计 实验四 桥接模式实验
spingboot 容器项目完成CICD部署
SQL(二) —— join窗口函数视图
Day1:用原生JS把你的设备变成一台架子鼓!
将照片形式的纸质公章转化为电子公章(不需要下载ps)
第二讲 Linear Model 线性模型
解决端口占用问题
随机推荐
day8字典作业
DOM and its applications
【过一下6】机器视觉视频 【过一下2被挤掉了】
The software design experiment four bridge model experiment
flink部署操作-flink standalone集群安装部署
el-table鼠标移入表格改变显示背景色
flink项目开发-配置jar依赖,连接器,类库
Flink Broadcast 广播变量
day12函数进阶作业
BFC(Block Formatting Context)
Xiaobai, you big bulls are lightly abused
数据库实验五 备份与恢复
el-pagination分页分页设置
spark-DataFrame数据插入mysql性能优化
DOM及其应用
Redux
学习总结week3_3迭代器_模块
vscode+pytorch使用经验记录(个人记录+不定时更新)
ES6基础语法
基于Flink CDC实现实时数据采集(四)-Sink接口实现