当前位置:网站首页>2020 Bioinformatics | TransformerCPI
2020 Bioinformatics | TransformerCPI
2022-07-04 04:21:00 【Stunned flounder (】
2020 Bioinformatics | TransformerCPI: Improving compound–protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments

Paper: https://academic.oup.com/bioinformatics/article/36/16/4406/5840724?login=false
Code: https://github.com/lifanchen-simm/transformerCPI
Abstract
Identifying compounds - Protein interactions (CPI) It is a key task in drug discovery and chemical genomics research , Proteins without three-dimensional structure account for a large part of potential biological targets , This requires the development of prediction using only protein sequence information CPI Methods . However , Sequence based CPI The model may face some specific defects , Including the use of inappropriate data sets 、 Hidden ligand bias and improper splitting of data sets , This leads to overestimation of its prediction performance .
result
To solve these problems , Here we have built a dedicated for CPI Predicted new data set , A new method named TransformerCPI New transformer neural network , A more rigorous label reversal experiment is introduced to test whether the model learns the real interaction characteristics .TransformerCPI Significantly improved performance has been achieved in the new experiment , And it can be deconvoluted to highlight the important interaction regions between protein sequences and compound atoms , This may contribute to the study of Chemical Biology , And provide useful guidance for further ligand structure optimization .
The current problems
- ** Use inappropriate data sets :** The learning of the model mainly depends on the data set it inputs , Inappropriate data sets make the model easy to deviate from the target . Based on chemical genomics CPI Modeling , The overall goal of modeling is to predict the interactions between different proteins and compounds based on the abstract representation of the characteristics of proteins and ligands . There is a collection of DUD-E Data sets , Designed to train structure based virtual screening . Besides ,DUD-E、MUV、Human and BindingDB Most of the ligands in appear in only one class , Negative samples are generated by algorithms that may introduce undetectable noise . These datasets can be separated by ligand information , And there is no guarantee that the model learns protein information or interaction characteristics .
- Hide ligand deviation :. Structure based virtual filtering 、 be based on 3D-CNN And others in DUD-E Training model on dataset (Sieg wait forsomeone ,2019 year )) It has been pointed out that prediction is mainly based on ligand modes rather than interaction characteristics , It leads to the mismatch between theoretical modeling and practical application .
- ** Improperly split data sets :** Machine learning researchers randomly divide the data into training sets and test sets . However , The traditional classification measurement is used on the randomly divided test set , It is not clear whether the model learns the real interaction features or other unexpected hidden variables , This may lead to accurate models that answer the wrong questions .
TransformerCPI The model architecture of
The model proposed by the author is based on transformer framework , The architecture was originally designed for neural machine translation tasks .transformer It is an automatic regression encoder - Decoder model , It combines multi head attention layer and position feedforward function to solve sequence to sequence tasks . Many pre training models are limited to seq2seq Mission , But the author was inspired by his powerful ability to capture features between two sequences , The converter structure was modified to predict compounds and proteins as two sequences CPI.TransformerCPI An overview of this is shown in the figure 2 Shown , Which retains transformer The decoder of , And modify its encoder and the final linear layer .
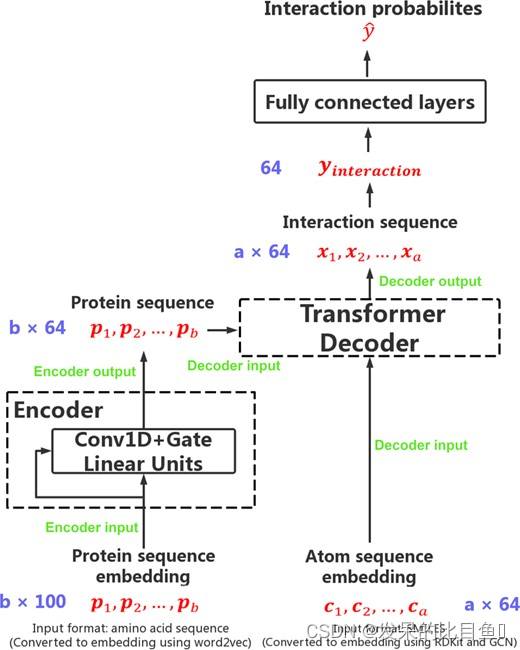
To convert protein sequences into sequential representations , The author first divided the protein sequence into overlapping 3-gram The amino acid sequence of , Then through the pre training method word2vec Translate all words into real values and embed .Word2vec It's an unsupervised Technology , It is used to learn high-quality distributed vector representations that describe complex syntactic and semantic word relationships . Integrate Skip-Gram and CBOW,word2vec Finally, words can be mapped to low dimensional real valued vectors , Among them, words with similar semantics map to vectors close to each other .
Then the sequence feature vector of the protein is passed to the encoder , To understand more abstract representations of proteins . The author replaces the original self attention layer in the encoder with a relatively simple structure . Considering the traditional transformer Architecture usually requires a huge training corpus , And it is easy to fit on small or medium-sized data sets , Therefore, the author uses a Conv1D And linear gated convolution network . Because it shows better performance on the data set designed by the author . The input of the gated convolution network is the protein eigenvector .
The author will design to solve the problem of semi supervised node classification GCN Move to solving the problem of molecular representation . When obtaining protein sequence representation and atomic representation . The interactive function is through transformer The decoder of , The decoder consists of a self attention layer and a feedforward layer . Protein sequence is the input of encoder , The atomic sequence is the input of the decoder , The output of the decoder is an interaction sequence with interaction characteristics and the same length as the atomic sequence . And the author modified the mask operation of the decoder , To ensure that the model can access the entire sequence , This is one of the most critical modifications to transform the transformation architecture from autoregressive tasks to classification tasks .
Last , Feed back the final interaction eigenvector to the following fully connected layers , And return the probability of interaction between compound and protein y.
Data sets
Open dataset : Three benchmark datasets , Human data sets , Caenorhabditis elegans data set and BindingDB Data sets .
** Label inversion dataset :** Many previous studies have passed CPI The random cross combination of pairs or the use of similarity based methods to generate negative samples , This may introduce unexpected noise and unnoticed deviations . First , The author from GLASS The database builds a GPCR Data sets . secondly , The author is based on KIBA The dataset builds Kinase(Kinase, Activating enzyme ) Data sets .
In order to confirm the interactive function of the actual learning of the model and accurately evaluate the impact of hidden variables , The author proposed a more rigorous label reversal experiment . The ligands in the training set only appear in one class of samples ( Positive or negative interaction CPI Yes ), Ligands only appear in another class of samples in the test set . In this way , Models are forced to use protein information to understand interaction patterns , And make opposite predictions for those selected ligands . If the model only stores ligand patterns , It is impossible to make a correct prediction , Because its stored ligand has the wrong ( contrary ) label . therefore , The label reversal experiment is specially designed to evaluate the results based on chemical genomics CPI Model , And it can indicate how much influence the hidden ligand bias has .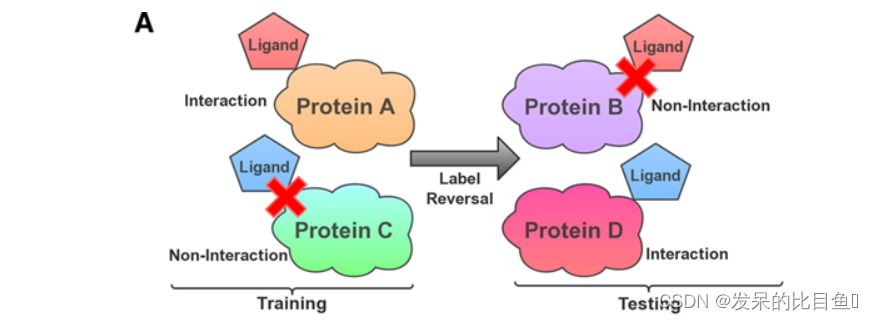
summary
The transformation architecture with self attention mechanism is modified to solve sequence based CPI Classification task , Thus, a system named TransformerCPI Model of , The model shows high performance on three benchmark datasets . To address the potential risks of deep learning , The authors constructed a specific genomics based on Chemistry CPI New dataset for the task , And designed a more rigorous label reversal experiment . Compared with other models , In the new experiment ,TransformerCPI Excellent performance , It shows that it can learn the required interaction characteristics and reduce the risk of hidden ligand offset . Last , The explanatory power of the model is studied by mapping attention weights to protein sequences and complex atoms , This can help the author determine whether the prediction is reliable and physically meaningful . Overall speaking ,TransformerCPI It provides a way to explain the model , It also provides useful guidance for further experimental optimization .
边栏推荐
- STM32外接DHT11显示温湿度
- Lnk2038 detected a mismatch of "runtimelibrary": the value "md_dynamicrelease" does not match the value "mdd_dynamicdebug" (in main.obj)
- Why is the probability of pod increasing after IPtable
- 毕业三年,远程半年 | 社区征文
- The three-year revenue is 3.531 billion, and this Jiangxi old watch is going to IPO
- 【CSRF-01】跨站请求伪造漏洞基础原理及攻防
- SDP中的SPA
- JS实现文字滚动 跑马灯效果
- Mitsubishi M70 macro variable reading Mitsubishi M80 public variable acquisition Mitsubishi CNC variable reading acquisition Mitsubishi CNC remote tool compensation Mitsubishi machine tool online tool
- STM32 external DHT11 display temperature and humidity
猜你喜欢
Idea modify body color

Confession code collection, who says program apes don't understand romance

ctf-pikachu-CSRF

96% of the collected traffic is prevented by bubble mart of cloud hosting
Brief explanation of depth first search (with basic questions)
ctf-pikachu-XSS

Flink学习6:编程模型
Pytest multi process / multi thread execution test case

PPt 教程,如何在 PowerPoint 中将演示文稿另存为 PDF 文件?
TCP-三次握手和四次挥手简单理解
随机推荐
'2'>' 10'==true? How does JS perform implicit type conversion?
Support the first triggered go ticker
一位毕业生的自我分享
postgresql 用户不能自己创建表格配置
Three years of graduation, half a year of distance | community essay solicitation
RHCSA 03 - 文件的基础权限
【CSRF-01】跨站请求伪造漏洞基础原理及攻防
LNK2038 检测到“RuntimeLibrary”的不匹配项: 值“MD_DynamicRelease”不匹配值“MDd_DynamicDebug”(main.obj 中)
Rhcsa-- day one
User defined path and file name of Baidu editor in laravel admin
RHCSA 08 - automount配置
Cesiumjs 2022^ source code interpretation [0] - article directory and source code engineering structure
Balance between picture performance of unity mobile game performance optimization spectrum and GPU pressure
Epidemic strikes -- Thinking about telecommuting | community essay solicitation
Tcpclientdemo for TCP protocol interaction
VIM add interval annotation correctly
Distributed system: what, why, how
Redis cluster uses Lua script. Lua script can also be used for different slots
[Huawei cloud IOT] reading notes, "Internet of things: core technology and security of the Internet of things", Chapter 3 (I)
There is a problem that the package cannot be parsed in the like project
