当前位置:网站首页>Cluster script of data warehouse project
Cluster script of data warehouse project
2022-07-05 05:29:00 【SYBY】
Catalog
- Previous e-commerce warehouse project , To sum up
- Synchronous distribution scripts
- hadoop Group start and stop script for
- zookeeper Group start and stop script for
- kafka Group start and stop script for
- User behavior log collection end Flume Group start and stop script
- User behavior log collection end Flume To configure
- User behavior log, group start and group stop script on the consumer side
- User behavior log consumer Flume To configure
- Business data collection increment table Flume To configure
- Use Maxwell Script for full synchronization of incremental table on the first day
- DataX Sync script
Previous e-commerce warehouse project , To sum up
Synchronous distribution scripts
#!/bin/bash
#1. Judge the number of parameters
if [ $# -lt 1 ]
then
echo Not Enough Arguement!
exit;
fi
#2. Traverse all machines in the cluster
for host in hadoop102 hadoop103 hadoop104
do
echo ==================== $host ====================
#3. Traverse all directories , Send... One by one
for file in [email protected]
do
#4. Judge whether the file exists
if [ -e $file ]
then
#5. Get parent directory
pdir=$(cd -P $(dirname $file); pwd)
#6. Get the name of the current file
fname=$(basename $file)
ssh $host "mkdir -p $pdir"
rsync -av $pdir/$fname $host:$pdir
else
echo $file does not exists!
fi
done
done
hadoop Group start and stop script for
#!/bin/bash
if [ $# -lt 1 ]
then
echo "No Args Input..."
exit ;
fi
case $1 in
"start")
echo " =================== start-up hadoop colony ==================="
echo " --------------- start-up hdfs ---------------"
ssh hadoop102 "/opt/module/hadoop-3.1.3/sbin/start-dfs.sh"
echo " --------------- start-up yarn ---------------"
ssh hadoop103 "/opt/module/hadoop-3.1.3/sbin/start-yarn.sh"
echo " --------------- start-up historyserver ---------------"
ssh hadoop102 "/opt/module/hadoop-3.1.3/bin/mapred --daemon start historyserver"
;;
"stop")
echo " =================== close hadoop colony ==================="
echo " --------------- close historyserver ---------------"
ssh hadoop102 "/opt/module/hadoop-3.1.3/bin/mapred --daemon stop historyserver"
echo " --------------- close yarn ---------------"
ssh hadoop103 "/opt/module/hadoop-3.1.3/sbin/stop-yarn.sh"
echo " --------------- close hdfs ---------------"
ssh hadoop102 "/opt/module/hadoop-3.1.3/sbin/stop-dfs.sh"
;;
*)
echo "Input Args Error..."
;;
esac
zookeeper Group start and stop script for
#!/bin/bash
case $1 in
"start"){
for i in hadoop102 hadoop103 hadoop104; do
echo "--------------zookeeper $i start-up --------------"
ssh $i "/opt/module/zookeeper-3.5.7/bin/zkServer.sh start"
done
};;
"stop"){
for i in hadoop102 hadoop103 hadoop104; do
echo "--------------zookeeper $i close --------------"
ssh $i "/opt/module/zookeeper-3.5.7/bin/zkServer.sh stop"
done
};;
"status"){
for i in hadoop102 hadoop103 hadoop104; do
echo "--------------zookeeper $i state --------------"
ssh $i "/opt/module/zookeeper-3.5.7/bin/zkServer.sh status"
done
};;
esac
kafka Group start and stop script for
#!/bin/bash
case $1 in
"start"){
for i in hadoop102 hadoop103 hadoop104; do
echo "--------- start-up $i Kafka---------"
ssh $i "/opt/module/kafka/bin/kafka-server-start.sh -daemon /opt/module/kafka/config/server.properties"
done
};;
"start"){
for i in hadoop102 hadoop103 hadoop104; do
echo "--------- start-up $i Kafka---------"
ssh $i "/opt/module/kafka/bin/kafka-server-stop.sh stop"
done
};;
esac
User behavior log collection end Flume Group start and stop script
#!/bin/bash
case $1 in
"start"){
for i in hadoop102 hadoop103
do
echo " -------- start-up $i collection flume-------"
ssh $i "nohup /opt/module/flume/bin/flume-ng agent -n a1 -c /opt/module/flume/conf/ -f /opt/module/flume/job/file_to_kafka.conf >/dev/null 2>&1 &"
done
};;
"stop"){
for i in hadoop102 hadoop103
do
echo " -------- stop it $i collection flume-------"
ssh $i "ps -ef | grep file_to_kafka.conf | grep -v grep |awk '{print \$2}' | xargs -n1 kill -9 "
done
};;
esac
User behavior log collection end Flume To configure
Flume Of source,channel and sink Use the following types :
1.taildir source 2.kafka channel 3. Not configured sink
# Defining components
a1.sources=r1
a1.channels=c1
# To configure source (taildirsource)
a1.sources.r1.type=TAILDIR
a1.sources.r1.filegroups=f1
a1.sources.r1.filegroups.f1=/opt/module/applog/log/app.*
a1.sources.r1.positionFile=/opt/module/flume/taildir_position.json
# Configure interceptors interceptors(ETL Data cleaning Judge json Is it complete )
a1.sources.r1.interceptors=i1
a1.sources.r1.interceptors.i1.type=com.serenity.flume.interceptor.ETLInterceptor$Builder
# To configure channel
a1.channels.c1.type=org.apache.flume.channel.kafka.KafkaChannel
a1.channels.c1.kafka.bootstrap.servers=hadoop102:9092,hadoop103:9092
a1.channels.c1.kafka.topic=topic_log
a1.channels.c1.parseAsFlumeEvent=false
# To configure sink( No, )
# Splice components
a1.sources.r1.channels=c1
User behavior log, group start and group stop script on the consumer side
#!/bin/bash
case $1 in
"start")
echo " -------- start-up hadoop104 Log data flume-------"
ssh hadoop104 "nohup /opt/module/flume/bin/flume-ng agent -n a1 -c /opt/module/flume/conf -f /opt/module/flume/job/kafka_to_hdfs.conf >/dev/null 2>&1 &"
;;
"stop")
echo " -------- stop it hadoop104 Log data flume-------"
ssh hadoop104 "ps -ef | grep kafka_to_hdfs.conf | grep -v grep |awk '{print \$2}' | xargs -n1 kill -9"
;;
esac
User behavior log consumer Flume To configure
Flume Of source,channel and sink Use the following types :
1.kafka source 2.file channel 3.hdfs sink
## Components
a1.sources=r1
a1.channels=c1
a1.sinks=k1
## source1
a1.sources.r1.type = org.apache.flume.source.kafka.KafkaSource
a1.sources.r1.batchSize = 5000
a1.sources.r1.batchDurationMillis = 2000
a1.sources.r1.kafka.bootstrap.servers = hadoop102:9092,hadoop103:9092,hadoop104:9092
a1.sources.r1.kafka.consumer.group.id = flume2
a1.sources.r1.kafka.topics=cart_info,comment_info,coupon_use,favor_info,order_detail,order_detail_activity,order_detail_coupon,order_info,order_refund_info,order_status_log,payment_info,refund_payment,user_info
a1.sources.r1.interceptors = i1
a1.sources.r1.interceptors.i1.type = com.serenity.flume.interceptor.TimestampInterceptor$Builder
## channel1
a1.channels.c1.type = file
a1.channels.c1.checkpointDir = /opt/module/flume/checkpoint/behavior2
a1.channels.c1.dataDirs = /opt/module/flume/data/behavior2/
a1.channels.c1.maxFileSize = 2146435071
a1.channels.c1.capacity = 1000000
a1.channels.c1.keep-alive = 6
## sink1
a1.sinks.k1.type = hdfs
a1.sinks.k1.hdfs.path = /origin_data/gmall/db/%{
topic}_inc/%Y-%m-%d
a1.sinks.k1.hdfs.filePrefix = db-
a1.sinks.k1.hdfs.rollInterval = 10
a1.sinks.k1.hdfs.rollSize = 134217728
a1.sinks.k1.hdfs.rollCount = 0
## The control output file is a native file .
a1.sinks.k1.hdfs.fileType = CompressedStream
a1.sinks.k1.hdfs.codeC = gzip
## assemble
a1.sources.r1.channels = c1
a1.sinks.k1.channel= c1
Business data collection increment table Flume To configure
The start and stop script is the same as the previous Flume The startup and shutdown scripts are basically the same , Pay attention to the configuration address .
Flume Of source,channel and sink Use the following types :
1.kafka source 2.file channel 3.hdfs sink
TimestampInterceptor And user behavior logs Flume Of TimestampInterceptor, Different time units .
## Components
a1.sources=r1
a1.channels=c1
a1.sinks=k1
## source1
a1.sources.r1.type = org.apache.flume.source.kafka.KafkaSource
a1.sources.r1.batchSize = 5000
a1.sources.r1.batchDurationMillis = 2000
a1.sources.r1.kafka.bootstrap.servers = hadoop102:9092,hadoop103:9092,hadoop104:9092
a1.sources.r1.kafka.consumer.group.id = flume2
a1.sources.r1.kafka.topics=cart_info,comment_info,coupon_use,favor_info,order_detail,order_detail_activity,order_detail_coupon,order_info,order_refund_info,order_status_log,payment_info,refund_payment,user_info
a1.sources.r1.interceptors = i1
a1.sources.r1.interceptors.i1.type = com.serenity.flume.interceptor.db.TimestampInterceptor$Builder
## channel1
a1.channels.c1.type = file
a1.channels.c1.checkpointDir = /opt/module/flume/checkpoint/behavior2
a1.channels.c1.dataDirs = /opt/module/flume/data/behavior2/
a1.channels.c1.maxFileSize = 2146435071
a1.channels.c1.capacity = 1000000
a1.channels.c1.keep-alive = 6
## sink1
a1.sinks.k1.type = hdfs
a1.sinks.k1.hdfs.path = /origin_data/gmall/db/%{
topic}_inc/%Y-%m-%d
a1.sinks.k1.hdfs.filePrefix = db-
a1.sinks.k1.hdfs.rollInterval = 10
a1.sinks.k1.hdfs.rollSize = 134217728
a1.sinks.k1.hdfs.rollCount = 0
## The control output file is a native file .
a1.sinks.k1.hdfs.fileType = CompressedStream
a1.sinks.k1.hdfs.codeC = gzip
## assemble
a1.sources.r1.channels = c1
a1.sinks.k1.channel= c1
a1.sinks.k1.hdfs.rollInterval = 10
a1.sinks.k1.hdfs.rollSize = 134217728
a1.sinks.k1.hdfs.rollCount = 0
Use Maxwell Script for full synchronization of incremental table on the first day
In the use of Maxwell After the first day full synchronization of the incremental table , Find data , Need to use where type='bootstrap-insert’ Filter , Because use bootstrap The first place for full synchronization will be marked with start and end marks ,type='bootstrap-start’ and type=‘bootstrap-complete’, It does not contain data .
#!/bin/bash
# The role of this script is to initialize all incremental tables , Only once
MAXWELL_HOME=/opt/module/maxwell
import_data() {
$MAXWELL_HOME/bin/maxwell-bootstrap --database gmall --table $1 --config $MAXWELL_HOME/config.properties
}
case $1 in
" Table name ")
import_data Table name
;;
.
.
.
" Table name ")
import_data Table name
;;
" Table name ")
import_data Table name
;;
"all")
# Synchronize multiple tables at once
import_data Table name
import_data Table name
import_data Table name
import_data Table name
import_data Table name
import_data Table name
import_data Table name
import_data Table name
import_data Table name
import_data Table name
;;
esac
DataX Sync script
# coding=utf-8
import json
import getopt
import os
import sys
import MySQLdb
#MySQL Related configuration , It needs to be modified according to the actual situation
mysql_host = "hadoop102"
mysql_port = "3306"
mysql_user = "root"
mysql_passwd = "000000"
#HDFS NameNode Related configuration , It needs to be modified according to the actual situation
hdfs_nn_host = "hadoop102"
hdfs_nn_port = "8020"
# The target path to generate the configuration file , It can be modified according to the actual situation
output_path = "/opt/module/datax/job/import"
# obtain mysql Connect
def get_connection():
return MySQLdb.connect(host=mysql_host, port=int(mysql_port), user=mysql_user, passwd=mysql_passwd)
# Get the metadata of the table Contains column names and data types
def get_mysql_meta(database, table):
connection = get_connection()
cursor = connection.cursor()
sql = "SELECT COLUMN_NAME,DATA_TYPE from information_schema.COLUMNS WHERE TABLE_SCHEMA=%s AND TABLE_NAME=%s ORDER BY ORDINAL_POSITION"
cursor.execute(sql, [database, table])
fetchall = cursor.fetchall()
cursor.close()
connection.close()
return fetchall
# obtain mysql The names of the tables
def get_mysql_columns(database, table):
return map(lambda x: x[0], get_mysql_meta(database, table))
# Add the obtained metadata to mysql The data type of is converted to hive Data type of Write to hdfswriter in
def get_hive_columns(database, table):
def type_mapping(mysql_type):
mappings = {
"bigint": "bigint",
"int": "bigint",
"smallint": "bigint",
"tinyint": "bigint",
"decimal": "string",
"double": "double",
"float": "float",
"binary": "string",
"char": "string",
"varchar": "string",
"datetime": "string",
"time": "string",
"timestamp": "string",
"date": "string",
"text": "string"
}
return mappings[mysql_type]
meta = get_mysql_meta(database, table)
return map(lambda x: {
"name": x[0], "type": type_mapping(x[1].lower())}, meta)
# Generate json file
def generate_json(source_database, source_table):
job = {
"job": {
"setting": {
"speed": {
"channel": 3
},
"errorLimit": {
"record": 0,
"percentage": 0.02
}
},
"content": [{
"reader": {
"name": "mysqlreader",
"parameter": {
"username": mysql_user,
"password": mysql_passwd,
"column": get_mysql_columns(source_database, source_table),
"splitPk": "",
"connection": [{
"table": [source_table],
"jdbcUrl": ["jdbc:mysql://" + mysql_host + ":" + mysql_port + "/" + source_database]
}]
}
},
"writer": {
"name": "hdfswriter",
"parameter": {
"defaultFS": "hdfs://" + hdfs_nn_host + ":" + hdfs_nn_port,
"fileType": "text",
"path": "${targetdir}",
"fileName": source_table,
"column": get_hive_columns(source_database, source_table),
"writeMode": "append",
"fieldDelimiter": "\t",
"compress": "gzip"
}
}
}]
}
}
if not os.path.exists(output_path):
os.makedirs(output_path)
with open(os.path.join(output_path, ".".join([source_database, source_table, "json"])), "w") as f:
json.dump(job, f)
def main(args):
source_database = ""
source_table = ""
options, arguments = getopt.getopt(args, '-d:-t:', ['sourcedb=', 'sourcetbl='])
for opt_name, opt_value in options:
if opt_name in ('-d', '--sourcedb'):
source_database = opt_value
if opt_name in ('-t', '--sourcetbl'):
source_table = opt_value
generate_json(source_database, source_table)
if __name__ == '__main__':
main(sys.argv[1:])
边栏推荐
- Binary search basis
- sync. Interpretation of mutex source code
- 剑指 Offer 09. 用两个栈实现队列
- [merge array] 88 merge two ordered arrays
- xftp7与xshell7下载(官网)
- [depth first search] 695 Maximum area of the island
- Animation scoring data analysis and visualization and it industry recruitment data analysis and visualization
- 剑指 Offer 04. 二维数组中的查找
- Introduction to memory layout of FVP and Juno platforms
- GBase数据库助力湾区数字金融发展
猜你喜欢
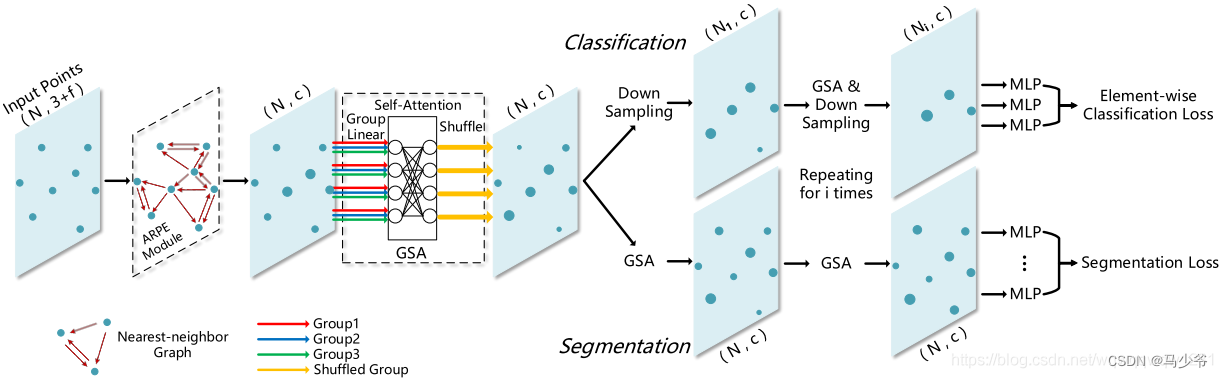
Improvement of pointnet++
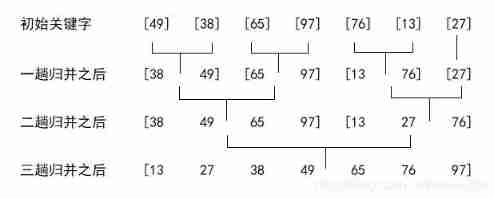
Merge sort
![To be continued] [UE4 notes] L4 object editing](/img/0f/cfe788f07423222f9eed90f4cece7d.jpg)
To be continued] [UE4 notes] L4 object editing
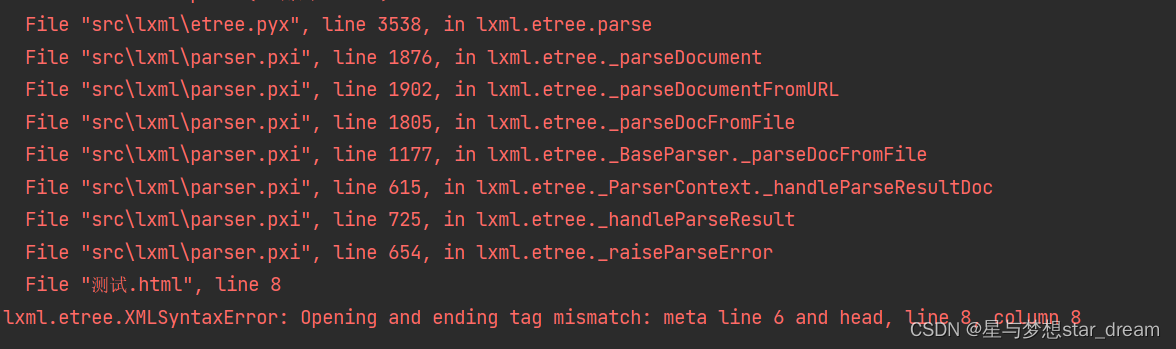
lxml. etree. XMLSyntaxError: Opening and ending tag mismatch: meta line 6 and head, line 8, column 8
一个新的微型ORM开源框架
YOLOv5-Shufflenetv2
Romance of programmers on Valentine's Day

Acwing 4300. Two operations
Reader writer model
![[转]: OSGI规范 深入浅出](/img/54/d73a8d3e375dfe430c2eca39617b9c.png)
[转]: OSGI规范 深入浅出
随机推荐
Acwing 4300. Two operations
剑指 Offer 05. 替换空格
发现一个很好的 Solon 框架试手的教学视频(Solon,轻量级应用开发框架)
动漫评分数据分析与可视化 与 IT行业招聘数据分析与可视化
[转]: OSGI规范 深入浅出
[binary search] 69 Square root of X
过拟合与正则化
Haut OJ 1243: simple mathematical problems
Haut OJ 1350: choice sends candy
Remote upgrade afraid of cutting beard? Explain FOTA safety upgrade in detail
Summary of Haut OJ 2021 freshman week
Acwing 4301. Truncated sequence
注解与反射
SSH password free login settings and use scripts to SSH login and execute instructions
Using HashMap to realize simple cache
Haut OJ 1357: lunch question (I) -- high precision multiplication
lxml. etree. XMLSyntaxError: Opening and ending tag mismatch: meta line 6 and head, line 8, column 8
26、 File system API (device sharing between applications; directory and file API)
一个新的微型ORM开源框架
剑指 Offer 09. 用两个栈实现队列