当前位置:网站首页>Imeta | Chen Chengjie / Xia Rui of South China Agricultural University released a simple method of constructing Circos map by tbtools
Imeta | Chen Chengjie / Xia Rui of South China Agricultural University released a simple method of constructing Circos map by tbtools
2022-07-07 00:04:00 【Liu Yongxin Adam】
Click on the blue words Pay attention to our
customized Circos chart : Use TBtools, From data preparation to visualization

https://onlinelibrary.wiley.com/doi/full/10.1002/imt2.35
●2022 year 7 month 4 Japan , Xia Rui team of South China Agricultural University is in iMeta Published online entitled “A painless way to customize Circos plot: From data preparation to visualization using TBtools” The article .
● In this article , The author in TBtools Developed in “Advanced Circos” function , Provide construction Circos A simple method of drawing .“Advanced Circos” Function provides a user-friendly interface , Used to customize parameter settings , It can also be used to visualize various genome level data , Such as genome related information 、 Comparison data 、 Gene density and QTL Location .
● First author : Chen Chengjie
● Corresponding author : Xia Rui ([email protected])
● joint authorship : Wuya
pick want
Circos Figure enables scientists to easily explore biological big data on a genome-wide scale , However, cumbersome input data preparation and complex parameter configuration limit its application . We are TBtools Developed in “Advanced Circos” function , Provide construction Circos A simple method of drawing . As a software out of the box ,TBtools It integrates a series of functions to facilitate the preparation of input data .“Advanced Circos” Function provides a user-friendly interface , Used to customize parameter settings , It can also be used to visualize various genome level data , Such as genome related information 、 Comparison data 、 Gene density and QTL Location . This paper introduces “Advance Circos” The main features and upstream data preparation methods , It aims to enable more users to use Circos Figure to explore genome big data .
key word :Circos, TBtools, Genome wide level , Data visualization
bright spot
● Out of the box solution for genome-wide data visualization
● Circos Figure drawing and parameter customization handle tutorial
● Reproducible project management
Video tutorial
Bilibili:https://www.bilibili.com/video/BV1iT411u7y1/
Youtube:https://youtu.be/ERWo7KhLQs0
Chinese translation 、PPT、 in / Download extended materials such as English video interpretation
Please visit the official website of the Journal :http://www.imeta.science/
Full text interpretation
Development motivation
With the rapid development of sequencing technology and the progress of data analysis technology , More and more biological genome sequences are decoded . With it , Biological research has entered the post genome era . This requires us to explore large-scale biological data more frequently from the whole genome .2009 year ,Krzywinski Put forward Circos Figure visualization mode . This is a powerful way to visualize genomic big data [1]. Since then ,Circos It has been used in many comparative genomic analyses , However, due to its complexity in drawing configuration and program use , Its utility has not yet been fully released . Although there are several methods for fast rendering Circos Tools for Graphs [1-7], But there are still places worth optimizing :(1) Most of the Circos The installation of drawing tools is complex , Or you need to work in a command-line environment , Users are required to have a high level of computer skills , Such as Perl-Circos;(2)Circos Graph drawing often requires users to use other kinds of software or write scripts to sort out data ;(3) Interactive editing is not supported for drawing drawings 、 The collaboration and sharing ability of direct redrawing and intermediate engineering files is limited .
therefore , We are TBtools [8] Developed in “Advanced Circos” function , Designed to provide the creation of Circos The simplest of the graph 、 The most convenient way . What users need to do is to follow TBtools Simple prompts in the graphical interface to organize tab delimited input files . All drawing parameters can be adjusted interactively ,Circos Graphs can be generated and refreshed immediately . Users can save work items for further modification 、 Copy and share . Besides ,TBtools As a multifunctional tool kit , It has a series of text processing and data processing functions , Help users prepare input data easily and quickly , by Circos Drawing creation provides a one-stop solution .
Data categories
TBtools Medium “Advanced Circos” The function supports visualization of multiple groups of continuous or discrete data . Usually , These data types can be divided into four categories , Pictured 1 Shown :
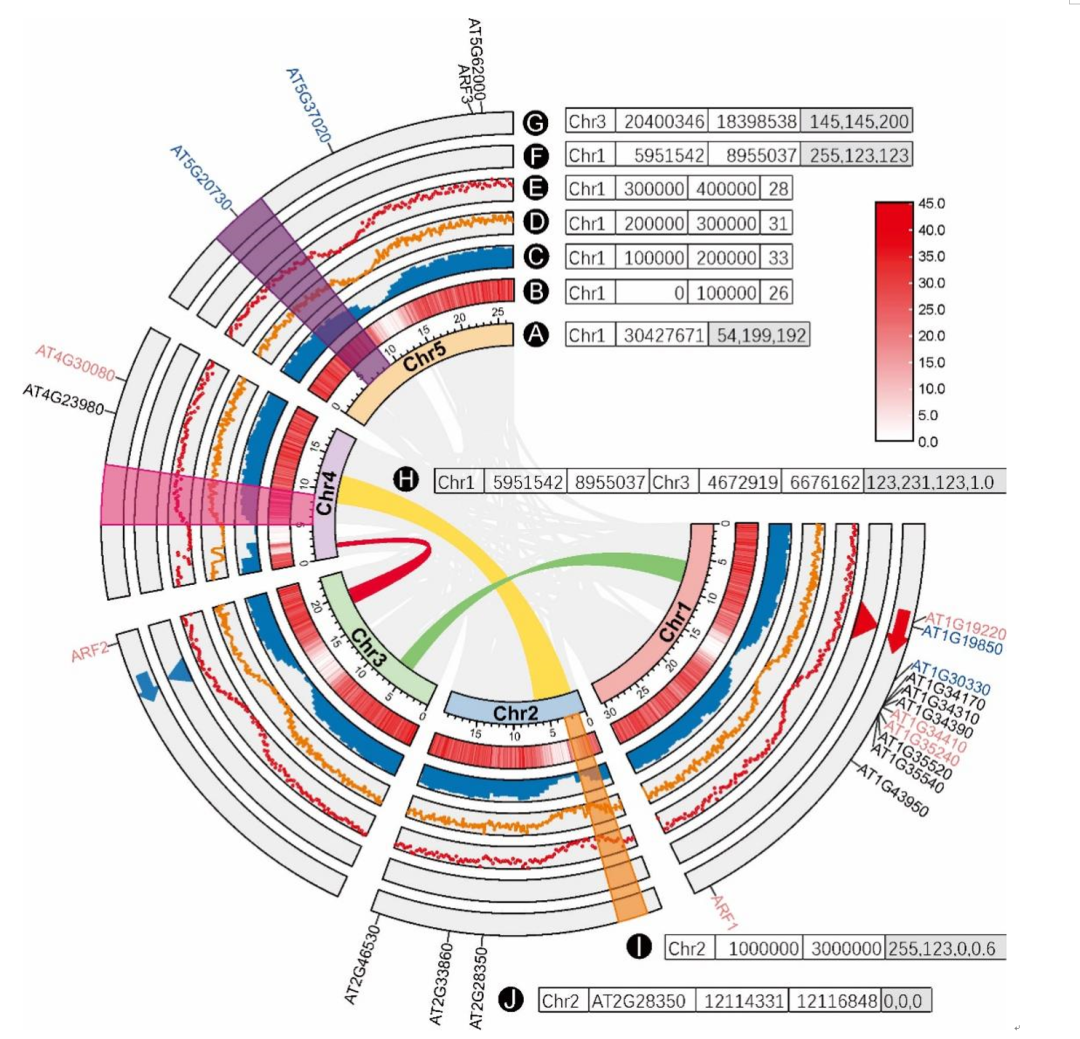
chart 1. Different data types are in TBtools Of “Advanced Circos” Visual mode of
(A) Chromosome skeleton ;(B) Thermogram ;(C) Bar chart ;(D) Broken line diagram ;(E) Point graph ;(F) triangle ;(G) arrow ;(H)bézier curve ;(I) block ;(J) Text label . Adjacent to each label character is an example of its corresponding input data format . Text with a gray background indicates that the corresponding column is optional .
The first is chromosome skeleton ( chart 1A), Present specific chromosomes or other genomic sequences ( Such as brackets or overlapping groups ). It is Circos The trunk of the diagram , Is the required input data . The default input data consists of two mandatory columns , One is chromosome ID, The other is chromosome length information . Users can also add options in the third column RGB Code , To specify the color of the chromosome skeleton .
The second is the marker data of specific chromosome regions ( chart 1J), It can be used to mark specific intervals , Such as genes or QTL( Quantitative trait loci ) Location . The corresponding input is a tab delimited file , It contains four mandatory columns and one optional column : Chromosome ID、 Area tag label 、 The starting coordinates 、 End coordinates and RGB Code ( Optional ).
The third type shows the association information of chromosome regions ( chart 1H), It is usually used to show homologous regions or chromosome interactions . Such data is usually placed in Circos The innermost layer of the graph . The input file consists of six mandatory columns and one optional column , Tab delimited , They are chromosomes ID、 The starting coordinates 、 Termination coordinates 、 Chromosome ID、 The starting coordinates 、 Termination coordinates and RGB Code ( Optional ).
The fourth category is chromosome region statistics ( chart 1B-G,1I), It can be displayed in various ways , Including continuous data ( Heat map 、 Bar chart 、 Line chart or dot chart display ) And discrete data ( In triangle 、 Arrow or tile / Rectangular display ). For continuous data , The format of the input file is “ Chromosome ID、 The starting coordinates 、 End coordinates and values indicated by numbers ”( chart 1B-E); For discrete data , The input file format is “ Chromosome ID、 The starting coordinates 、 End coordinates and RGB Code ”( chart 1F、1G、1I). For arrow symbols ( chart 1G), When the start coordinate is greater than the end coordinate , In the opposite direction . By adjusting the drawing span , Can be stacked Circos Different tracks on the drawing , To realize the combination of various visual types , Pictured 1I Sum graph 6 Shown . rectangular / Tile tracks are used to highlight some areas of chromosomes .
The graphical interface
● main interface
open TBtools->“Graphics”->“Advanced Circos”, In the pop-up interface ( chart 2A-E) in , There are three input file fields and two function buttons .
1、Chr Length file ( Required );
2、 Genomic characteristic information ( Optional );
3、 Associated area information ( Optional );
4. “Load Project…” Button loads pre-existing Circos project ;
5、“Show My Circos Plot!” Button to generate genome skeleton map .
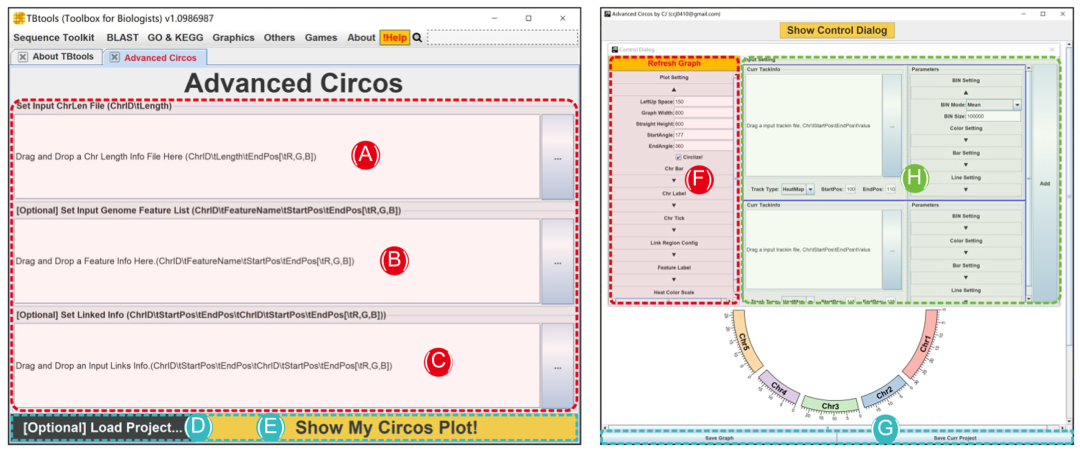
chart 2. Advanced Circos Main interface and parameter panel
(A)-(C) Input file panel ;(D) Existing item restore button ;(E) Click the button to generate a genome skeleton map ;(F)Cirocs Global parameter settings of the drawing ;(G) Buttons for exporting drawings or saving projects ;(H) Parameter settings for statistical visualization of each chromosome region .
● Advanced parameter settings
By clicking “Show Control Dialog” Button , You can open advanced Circos Control panel . The parameter setting panel can be divided into three main parts ( chart 2F-G).
1、 The Save button at the bottom of the interface ( chart 2G).“Save Graph” Button is used to export the current drawing , Support bitmap and vector formats ;“Save Curr. Project” Button can be used to save the project in progress , The project can pass the above “Load Project…” recovery , It can also be shared directly with other users or reproduced directly on other devices ;
2、 Global parameter settings on the left side of the interface ( chart 2F). These settings are used to control the overall drawing details , It is divided into several parts in detail : Graphic settings 、 Color bar 、 Color label 、 Color marks 、 The correlation information 、 Feature tags 、 Heat map color scale . And the top “Refresh Graph” The button is used to apply the adjusted parameters and refresh the current drawing .
3、 The single channel graphic control parameters located on the right side of the interface ( chart 2H). They are used to control the visual details of chromosome region statistics , Chromosome region statistics are also divided into several parts , Include BIN Set up 、 color setting 、Bar Settings and line settings . single click “ add to ” Button , The graphic track will be generated . For the detailed effect of each parameter , It is recommended that users use sample data to explore by themselves ( See supporting data 1).
● Case presentation
Advanced Circos yes TBtools One of the built-in features of , It is integrated with a series of functions that can be used to prepare input data . In this section , We will demonstrate step by step how to use TBtools Of Advanced Circos The function starts from the common biological big data , To Circos Graph generation .
(a) Chromosome skeleton file preparation
The chromosome skeleton is Circos The trunk of the diagram . have access to TBtools Medium “Fasta Stat” Function to obtain information about the genome from the genome sequence file ( chart S1).
Functional location :“Sequence Toolkit”->“Fasta Tools”->“Fasta Stats”;
Input file :“Arabidopsis_ thaliana.genome.fna”
The output file :“Arabidopsis_ thaliana.genome.fna.ChrLen.txt”
Operation steps :
1、 go to “Fasta Stats”;
2、 Set input file and output file ;
3、 Check “ Only keep the sequence length ”;
4、 Click on “ Start ” Button , The required information will be saved to the output file
(“Arabidopsis_thaliana.genome.fna.ChrLen.txt”).
notes : The file name of the presentation data is italicized
The output file (“Arabidopsis_thaliana.genome.fna.ChrLen.txt”) Contains the sequence length information of each chromosome . You can edit it manually . for example , Here we delete the length information of two plasmid chromosomes .
Chr1 30427671
Chr2 19698289
Chr3 23459830
Chr4 18585056
Chr5 26975502
ChrM 366924
ChrC 154478
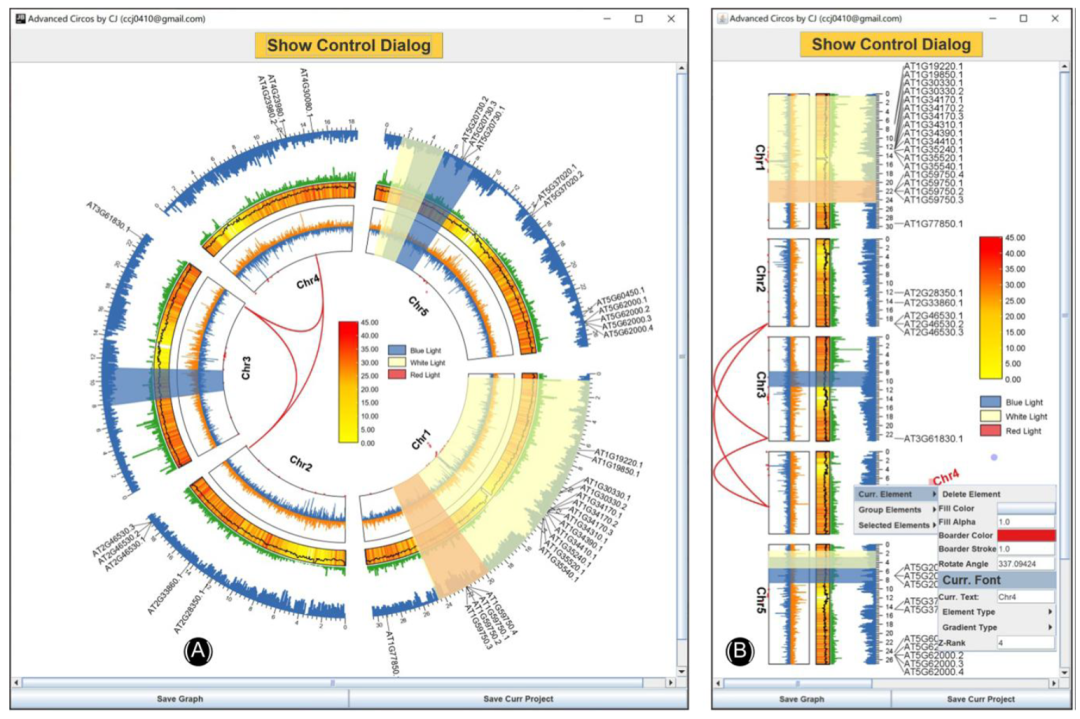
Just drag and drop the result file of chromosome length information to Advanced Circos( chart 2A) In the first field of , Press “Show My Circos Plot!” Button , Immediately generate the chromosome skeleton Circos chart ( chart 3A).
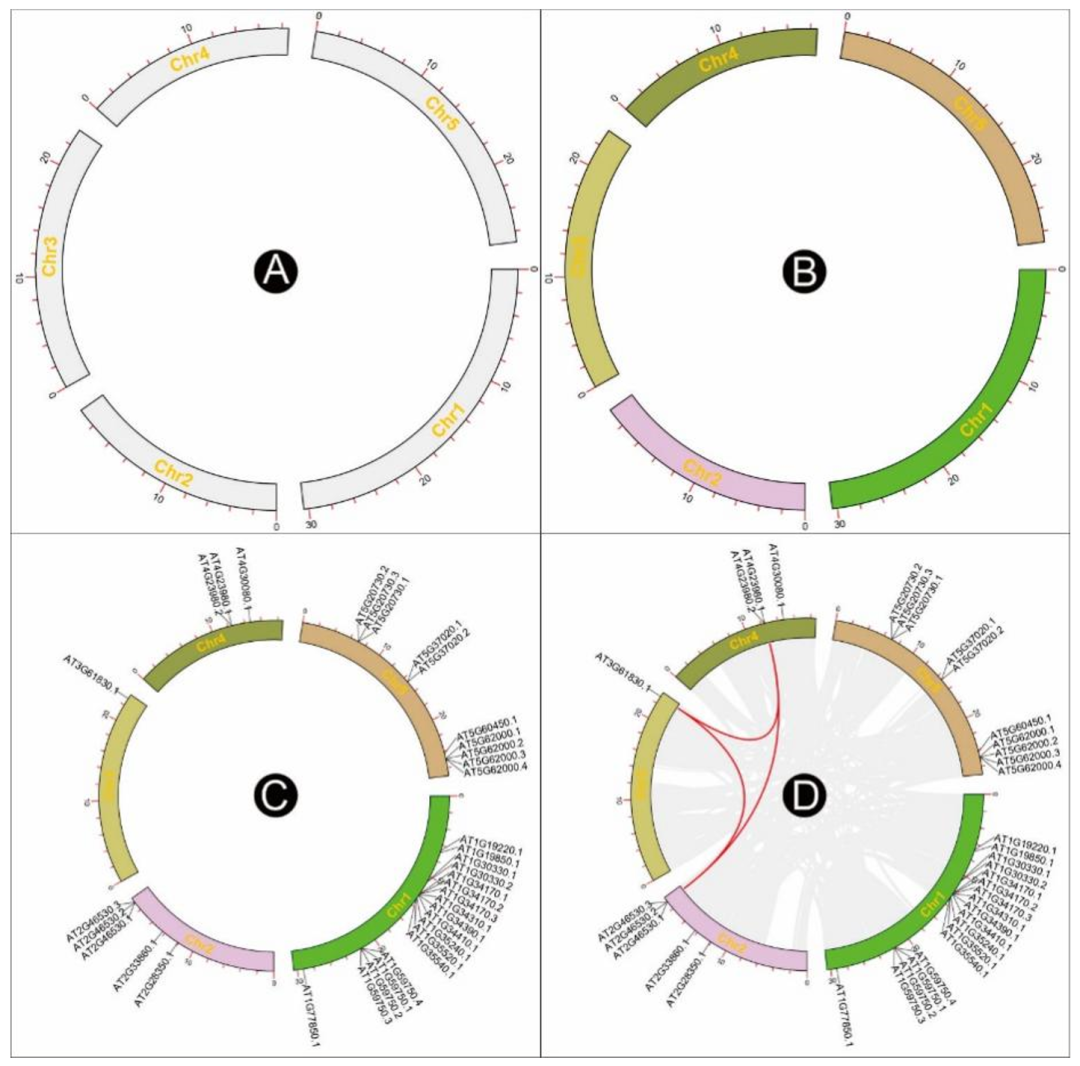
chart 3:Advanced Circos Chromosome skeleton 、 Visualization of area markers and area related data
(A) Basic chromosome skeleton ;(B) Chromosomal skeletons colored in different colors ;(C) Visualization of gene positions on the chromosome skeleton ;(D) Visualization of chromosome region association information between chromosomes .
By adding RGB Color code , Each chromosome can be colored differently . Users can also use “Discrete Color Scheme Generator” Automatically generate a series of colors ( chart S2)
Functional location :“ graphics ”->“ palette ”->“ Discrete color scheme generator ”;
Input file :“Arabidopsis_thaliana.genome.fna.ChrLen.txt”
The output file :“Arabidopsis_thaliana.genome.fna.ChrLen.withColor.txt”
Operation steps :
1、 Switch to “ Discrete color scheme generator ”;
2、 Set input file and output file ;
3、 Click on “ Start ” Button ,RGB The color code will be added to the output file (“Arabidopsis_thaliana.genome.fna.ChrLen.withColor.txt”).
The contents of the output file are as follows :
Chr1 30427671 11,249,4
Chr2 19698289 241,193,242
Chr3 23459830 208,201,119
Chr4 18585056 152,162,81
Chr5 26975502 212,178,129
stay Advanced Circos Replace the chromosome skeleton information with this file , A color skeleton diagram will be generated ( chart 3B).
(b) Add genome tag information
in many instances , We hope to mark some genomic features on chromosomes , For example, genes 、QTL and TAD( Topological association domain ). for example , To highlight certain genes , Users need to get the location information of each gene first .TBtools Of “GXF Gene location and information extraction ” Functions can be used to obtain the corresponding genomic regions of specific genes ( chart S3).
Functional location :“Sequence Toolkit” -> “GFF3/GTF Manipulate” -> “GXF Gene Position & Info Extract”
Input file :Arabidopsis_thaliana.genome.gff3
The output file :Arabidopsis_thaliana.genome.gff3.CDS.Info.xls
Operation steps :
1、 Set input 、 The output file
2、 Click on “Start” Button
The output file contains all of Arabidopsis CDS Location information for . Users can use Excel Or other text editing software to select the gene information of interest from the list . ad locum , We use TBtools Medium “Table Row Manipulate” function , Picked out Arabidopsis ARF Family genes ( Must be prepared in advance ARF Genetic ID list ).
Functional location :“Others” -> “Table/Text File Manipulator” -> “Table Row Manipulate”
Input file : Arabidopsis_thaliana.genome.gff3.CDS.Info.xls
Input ID list : ARF.Family.IDs.txt
The output file : Arabidopsis_thaliana.genome.ARF.Pos.Info.xls
Operation steps :
1、 Set input file , Select the first column to process ;
2、 Set input AtARF ID list ;
3、 Set output file ;
4、 Click on “Start” Button .
You can simply modify the result file , That is, only the first four columns are retained , The specific document information is as follows :
Chr1 AT1G19220.1 6628068 6633087
Chr1 AT1G19850.1 6886879 6891374
Chr1 AT1G30330.1 10685822 10690838
...
This file can be used directly Advanced Circos visualization . Users can also attach RGB Code , To render different text colors . If feature labels overlap in the drawing , You can adjust “ Global parameter setting ” in “ Feature tags ” Under the “ Overlapping weights ” value , If set to “-4” Or more , To optimize text spacing ( chart 3C)
(c) Genome region Association map
Chromosome sequences are interrelated within or between chromosomes , For example, sequence homology 、 Chromosome interaction and gene regulation .Circos Graphs are often used to show genome-wide replication 、 Genomic characteristics of large fragment translocation and tandem repeats . Users only need to use TBtools Some of the functions in , You can prepare relevant input files , for example ,“One Step MCScanX”[9]( chart S3).
Functional location :“Graphics” -> “Comparative Genomics” -> “One Step MCScanX”
Input genome sequence information :Arabidopsis_thaliana.genome.fna
Input gene structure annotation information : Arabidopsis_thaliana.genome.gff3
Output folder : Synteny Result
In the output directory , Will generate a suffix “*.geneLinkedRegion.tab.xls” The file of . It can be used for Advanced Circos visualization . The last column of the file ( The first 8 Column ) Contains information about homologous gene pairs ,Advanced Circos This information will be ignored . For all that , The user can choose the association interval of interest , adjustment RGB Code , And move these lines to the head of the input file to highlight these areas . Sample file , As shown below . The resulting graph will be similar to graph 3D.
Chr2 19105112 19108331 Chr3 22887889 22891435 255,0,0 AT2G46530.3.match.AT3G61830.1
Chr2 19105112 19108331 Chr4 12451277 12455014 255,0,0 AT2G46530.3.match.AT4G23980.1
Chr3 22887889 22891435 Chr4 12451277 12455014 255,0,0 AT3G61830.1.match.AT4G23980.1
……
(d) Genome data visualization
GC content / GCskew / Unknown base
Nucleic acid composition is a basic feature of the genome . for example ,GC Content and coding genes and functions DNA The density of elements is related ;GC-skew It is an index to measure guanine and cytosine chain preference , It can help detect in bacterial circular chromosomes DNA Replication start site . Unknown base (N) Distribution can show the quality of genome assembly to some extent .TBtools Medium “Fasta Window Stat” The function can be used to quickly transfer the... In the genome sequence file GC content 、GC Skew and N Tabulate the ratios .
Functional location :“Sequence Toolkit”->“Fasta Tools”->“Fasta Window Stat”
Input genome sequence file :Arabidopsis_thaliana.genome.fna
Output file prefix :Arabidopsis_thaliana.genome.Window.Stat
Three files with the same prefix will be generated . They are
“Arabidopsis_thaliana.genome.Window.Stat.GCratio”, “Arabidopsis_thaliana.genome.Window.Stat.GCskew”, “Arabidopsis_thaliana.genome.Window.Stat.Nratio”
These three output files are applicable to Advanced Circos visualization . here , We use N Ratio statistics file as the first example ( chart 4A).
1、 Delete manually N Than for 0 The line of ( Optional );
2、 After loading the chromosome skeleton information as described above , Click on “Start” Visualizing ;
3、 Enter the parameter control interface (“Show Control Dialog”), Click on the right “Add” Button , Add a new track ;
4、 Set up N Input file of ratio file ;
5、 Select the track type as the point , And then click “Refresh Graph”( chart 4B).
Please note that , We have used the sliding window method to calculate N ratio , therefore “BIN Mode” It can be set to “None”.
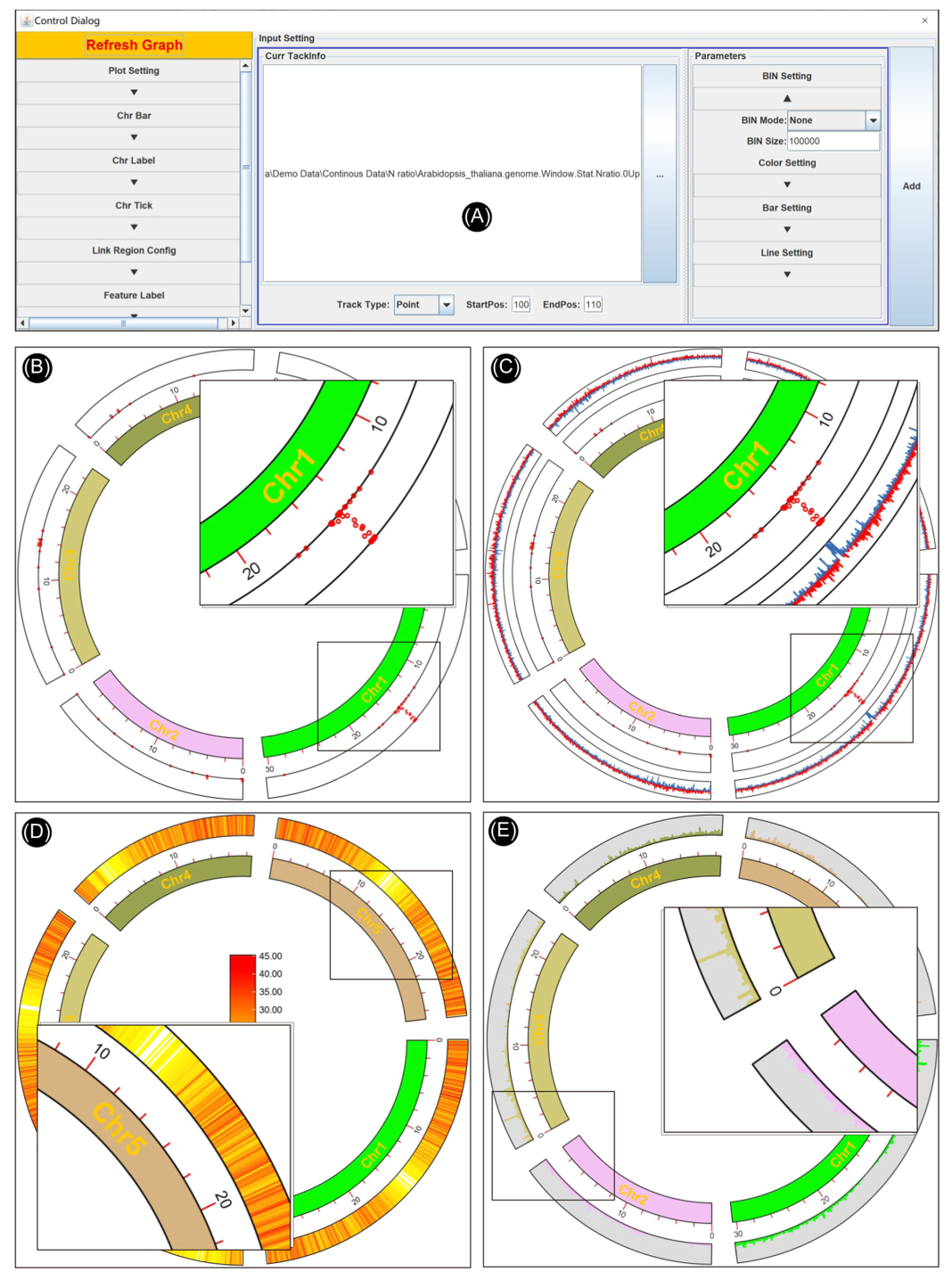
chart 4: Use different track types to view continuous data
(A) Track parameter panel ;(B) Viewed in the dot graph N Ratio ;(C)GC Skewness ;(D) Gene density in the heat map ;(E) Sort coverage in bar chart .
Similarly , We can use line graph to visualize GC Deflection .GC The offset is calculated in 0 As a boundary , Therefore, we will set “Sep Line Value” Set to 0, To achieve different shading of positive and negative offset values ( chart 4C).
Gene density
Genome wide gene density distribution is usually achieved by Circos Graph for visualization .TBtools There is a convenient function ——“Gene Density Profile”, It allows users to calculate gene density from gene structure annotation files , Usually it is GFF3 and GTF Format .
Functional location : “Sequence Toolkits” -> “GFF3/GTF Manipulate” -> “Gene Density Profile”
Input file : Arabidopsis_thaliana.genome.gff3
The output file : GeneDensity.profile
stay “Control Dialog” The palette , single click “Add” To get additional tracks , Drag and drop the gene density information file (“GeneDensity.profile”), Select heat map mode , Then refresh the drawing ( chart 4D). If the gene density distribution is obviously biased , The user can see “Heat Color Scale” Try different color scaling modes in the menu , For a better view . Each heat map track will automatically trigger the drawing of color scheme legend , And these legends can be easily moved .
Sequencing data coverage distribution
in many instances , In addition to the characteristics of the genome sequence itself , We are also interested in looking at the actual NGS Interested in the distribution of data on the genome , for example , Coverage of deep sequencing data .TBtools Medium “SAM/BAM/CRAM BIN Cov” The function can be used for the original alignment file generated by the mapping tool ( Usually it is SAM、BAM or CRAM Format ) Prepare input file .
Functional location : “Others” -> “HTS Data” -> “SAM/BAM/CRAM BIN Cov”
Input file : SRR17382349_1.bam
The output file : SRR17382349_1.bam.BINStat.tab.xls
here , We chose bar charts to visualize coverage data . Check “Color by Chr”, Make the color of the bar graph consistent with the color of the chromosome .“Bar Fill” Option controls the background color of the track , Set to gray here ( chart 4E).
Genomic variation data
Compare with the file ( For sequence coverage ) similar , stay TBtools in , Users can also process genomic variation data , for example , Containing genomic sequence variation information VCF file .“VCF BIN Cov” The function can be used for input preparation .
Functional location : “Others” -> “HTS Data” -> “VCF BIN Cov”
Input file : Est-1.qual50.filtered.direct.vcf.gz
The output file : Est-1.qual50.filtered.direct.vcf.BINCount.tab.xls
Use heat map mode , You can comprehensively view the hot spots of sequence variation on chromosomes . The overall pattern is largely complementary to the above gene density distribution .
QTLs/Arrow/TAD
All the demonstration data used above are continuous genome data ,“Advanced Circos” Various modes for displaying discrete data are also provided . We can refer to the above Tile track The format requires organization QTL Information , for example :
Chr3 7842037 11192039 56,108,176,0.7 Blue Light
Chr5 2899846 8562462 56,108,176,0.7 Blue Light
Chr1 19672910 24443023 227,26,28,0.7 Red Light
Chr1 592464 24581765 255,255,179,0.7 White Light
Chr5 1507102 5275918 255,255,179,0.7 White Light
Use this data as input , And select “Tile” Mode to display it ( chart 5A). Please note that , The last column is optional , Its existence will automatically trigger the legend generation . All legends can be easily moved . Besides ,“Advanced Circos” It can also be used for visualization TAD( Use “ triangle ” Pattern ) Or band direction gene interval ( Use “ arrow ” Pattern )( chart 1).
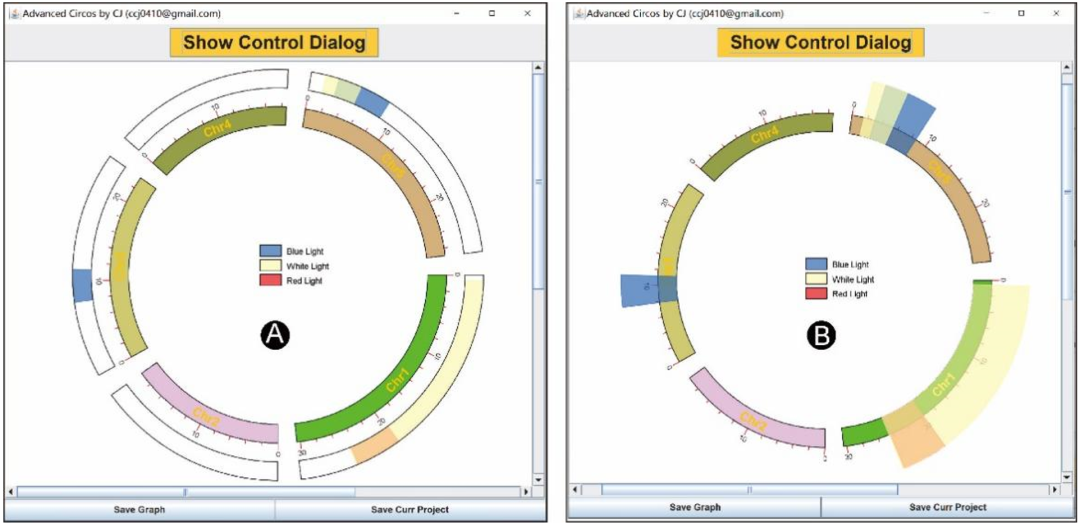
chart 5: Discrete data represented by different orbital types
(A)QTL Tile of ;(B) Modify the tile to highlight the genome spacing of interest .
(e) Other customization options
Highlight the section
Through adjustment Tile The drawing range of the track (“ tile ” Track “Start Pos” and “End Pos”), It can realize the function of highlighting areas . From the picture 5A Start , take “Start Pos” Set to 90,“Bar Border” Set to “null”, And refresh the image , We will get as shown in the figure 5B The picture shown .
Track overlap
Follow the same logic as the area highlighting function , You can combine different tracks into one , This can also be done by adjusting the “Start Pos” and “End Pos” To achieve . By simply merging the above tracks , We can get as shown in the figure 6A.
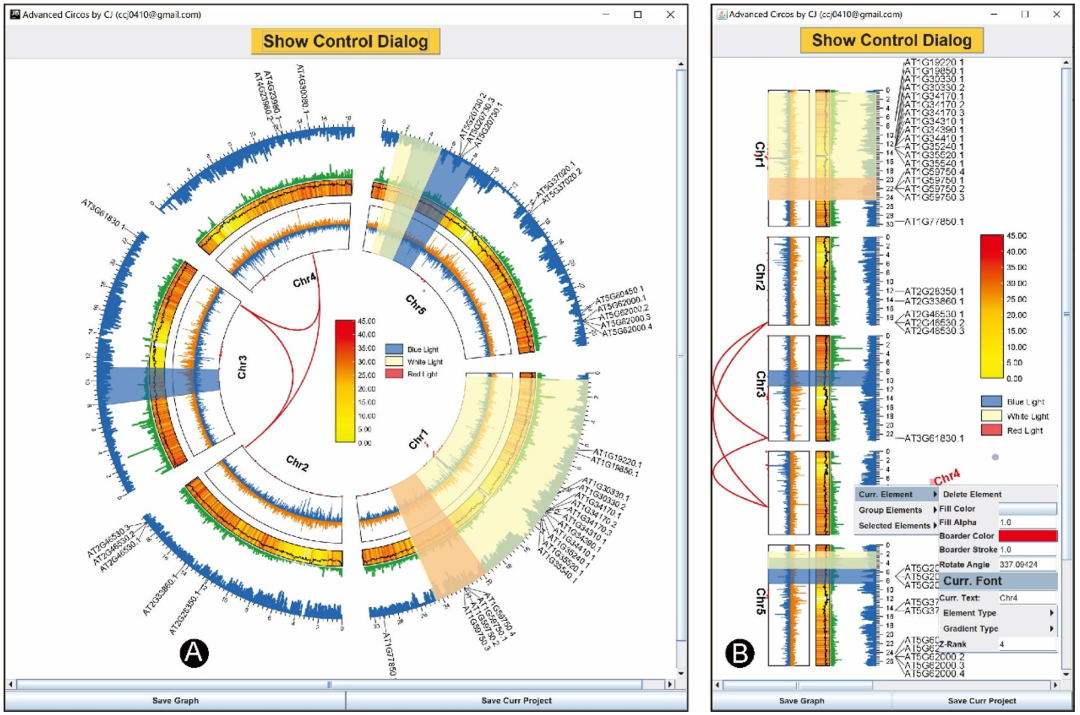
chart 6:Circos Overlapping tracks
(A) from Circos The inside and outside of the graph are : Genome region related Bézier Surfaces ;N- Dot graph of ratio distribution ;GCskew The line diagram of ; Heat map of gene density distribution and GC The line graph of ratio change overlaps ; Sequencing coverage bar graph ; Inverted bar chart ; Label of gene family ;(B) Worthy Circos
Linear or circular ?
Advanced Circos It is based on what we have been developing TBtools Powerful interactive drawing engine “JIGplot” Developed . therefore ,“Advanced Circos” With coordinate switching ( Between Cartesian coordinates and polar coordinates ) And interactive analysis . By deselecting “Circulized” Check box , We'll get a good value Circos chart , namely “Straight” Pattern ( chart 6B). Besides , Users can easily edit every element in the drawing , for example , Rotate elements and change text font and color .
preservation 、 Share and reload projects
In the main drawing panel , single click “Save Curr. Project”, Save the current drawing data and state to the specified directory . Users can directly from Advanced Circos The main interface reproduces the saved items , Make further adjustments . Besides , Users just need to package the directory and send it to colleagues to share their Circos project , Or work directly from other equipment .
Conclusion
Tedious data preparation 、 Complex configuration and a large number of text collation operations limit Circos The use of graphs in scientific research . This article briefly introduces TBtools in “Advanced Circos” Drawing function and corresponding parameter interface , And elaborated how from the common NGS Data departure , Use “Advanced Circos” Make informative Circos chart . Almost all steps can be done in TBtools It is easy to complete with a simple click . We expect , In this paper TBtools Of “Advanced Circos” It will enable more researchers to enjoy Circos Figure advantages in exploring large biological data .
thank
This study was conducted by Guangdong Laboratory of Lingnan Modern Agriculture ( Item number :NZ2021007)、 NSFC (32102320) And the special support plan of Guangdong Province (2019TX05N193) funding . We thank Dr. Yu haopeng of West China Hospital and Dr. Feng Junting of the Chinese Academy of Tropical Agricultural Sciences for their improvement TBtools Of Advanced Circos Constructive suggestions put forward by the function . We thank you very much 40000 A number of TBtools user , In especial 30 Good suggestions from several advanced users .
Conflict of interest
The author declares that there are no competing interests
Author's contribution
Chengjie Chen and Rui Xia Conceived the project ;Chengjie Chen and Rui Xia Designed the functions of the toolbox ;Chengjie Chen Performed all Java code .Ya Wu Tested these functions , And helped write tutorial manuals and charts .Chengjie Chen and Rui Xia Designed these figures and wrote the manuscript . All authors have read and passed the final manuscript .
Data availability
All demo data and TBtools The corresponding version of can be found in https://tbtools.cowtransfer.com/s/c60a5cfec3274f. Supplementary materials ( chart 、 surface 、 Script 、 Graphic abstract 、 Slide 、 video 、 Chinese translation and updated materials ) Available online DOI or iMeta Science Find http://www.imeta.science/.
a citation
Chen, Chengjie, Ya Wu, and Rui Xia. 2022. “A painless way to customize Circos plot: From data preparation to visualization using TBtools.” iMeta. e35. https://doi.org/10.1002/imt2.35
Author's brief introduction

Chen Chengjie ( First author )
● Teacher of horticulture College of South China Agricultural University , Shengxin tools TBtools author
● Now in Molecular Plant、Nature Communications、New Phytologist、Horticulture Research And other internationally renowned magazines 10 Yu Wen

Xia Rui ( Corresponding author )
● With concurrent , Professor, College of horticulture, South China Agricultural University
● At present, he is the academic backbone and young discipline leader of the State Key Laboratory for the protection and utilization of subtropical agricultural biological resources and the Key Laboratory for Horticultural Crop Biology and Germplasm Creation in South China of the Ministry of agriculture .2019 In, it was supported by the science and technology innovation leading talent project of the special support plan of Guangdong Province . Long term commitment to horticultural plant genome and small molecules RNA Related research , It has achieved internationally leading research results , stay Nature Genetics, Nature Communication, Molecular Plant and The Plant Cell And other academic publications SCI The paper 50 Multiple articles , Cumulative reference >5000 Time . At present, bioinformatics is mainly used 、 Genomics and Molecular Biology , The research focuses on biological issues such as the mechanism of flower sex differentiation of saparicaceae plants and the regulation mechanism of flower and fruit development of major southern fruits . And has developed a series of biological information data analysis tool database , Such as TBtools and sRNAanno(www.plantsRNAs.org) etc.
More recommendations
(▼ Click the jump )
iMeta Chinese translation of articles + Video interpretation
iMeta cover | One stop tool set for macro proteomics analysis iMetaLab Suite( University of Ottawa, Canada Figeys Group )
▸▸▸▸
iMeta | Wang Zhang group of South China Normal University reviewed the secret relationship between human lung microbiome and human health and disease
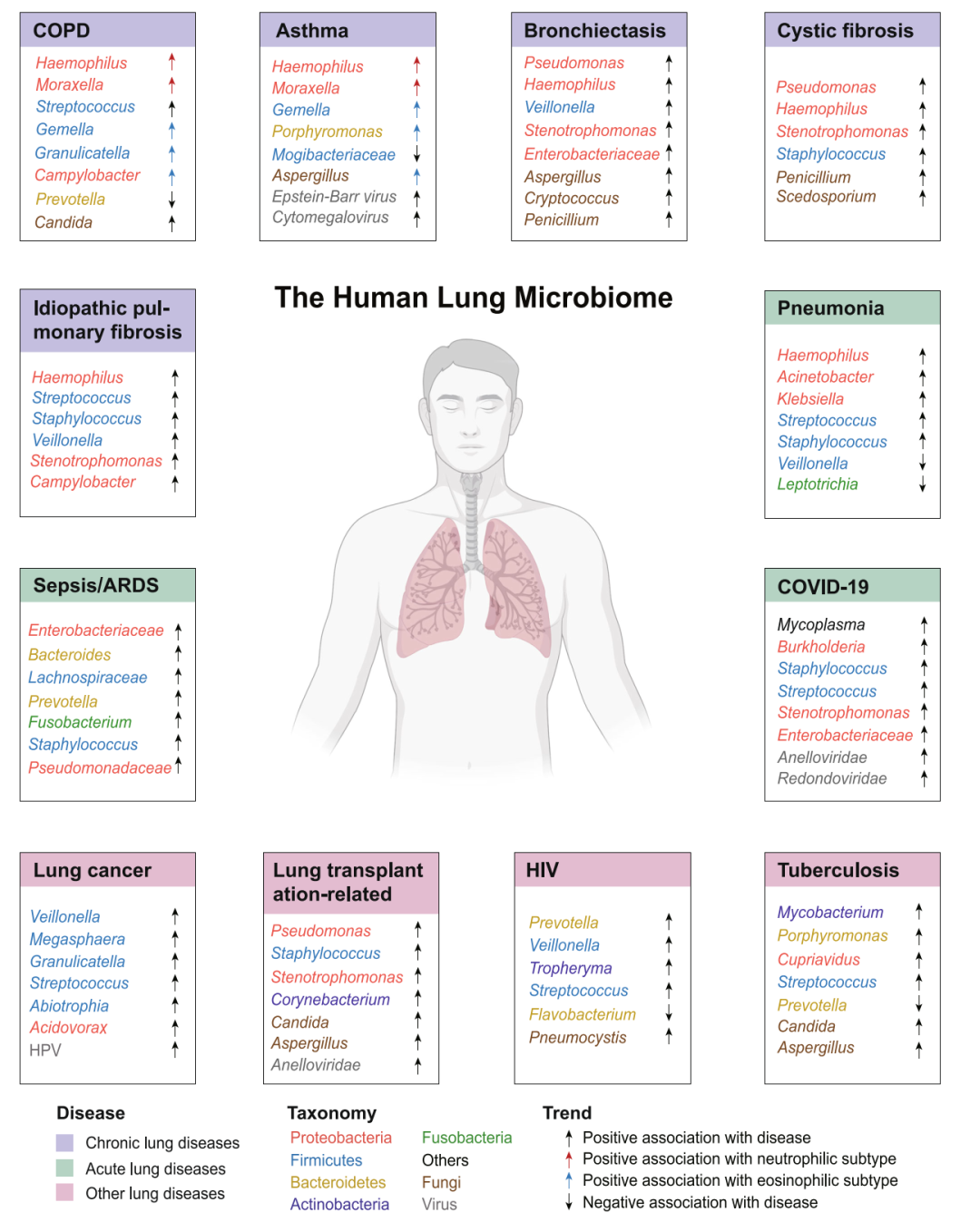
▸▸▸▸
iMeta | Nanopore sequencing of Xiayu formation of South University of science and technology reveals that microorganisms can reduce greenhouse gas emissions from high-altitude frozen soil
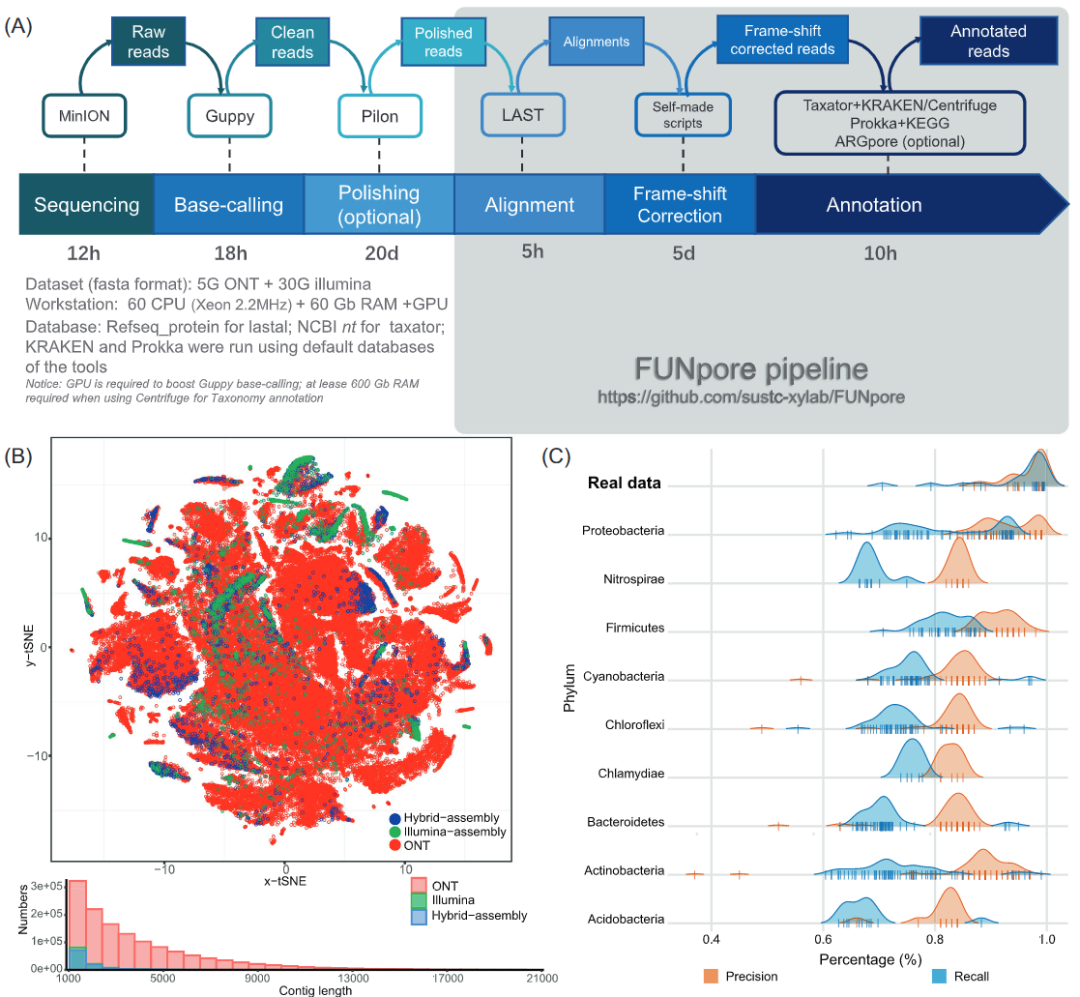
▸▸▸▸
iMeta | Chen Feng of Peking University / Chenzhibin et al. Published the experimental method of sampling each part in the study of oral microbiome (Protocol)
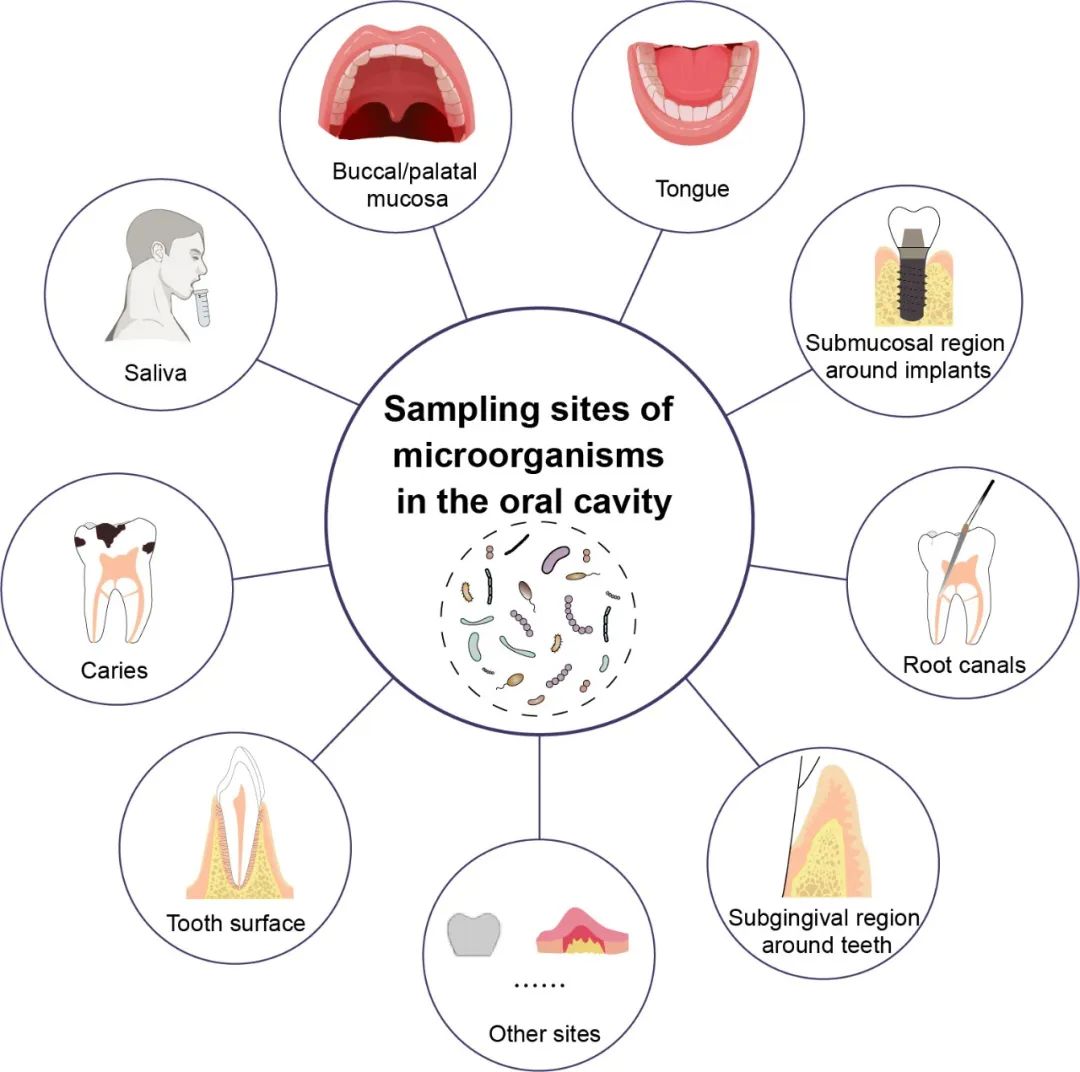
▸▸▸▸
iMeta | Zeng Zhenling of South China Agricultural University / Xiongwenguang et al - Study on the coexistence of drug resistance genes between pet dogs and their owners in the family
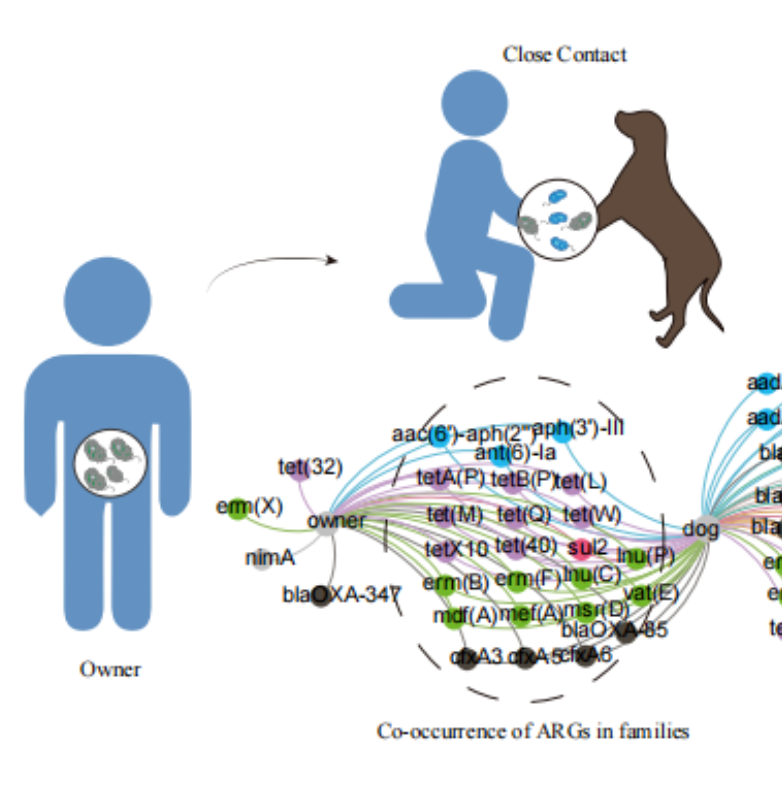
▸▸▸▸
iMeta | Mayingfei group of Shenzhen Advanced Research Institute has developed a method for analyzing intestinal flora based on neural network
▸▸▸▸
iMeta | Chenlianmin, et al. Of Nanjing Medical University summarized the potential impact of Enterobacteriaceae on complex diseases from the perspective of genome function
▸▸▸▸
iMeta | A review of CHENFENG group in Peking University standardization of oral microbiome : From technology driven to hypothesis driven
▸▸▸▸
iMeta | Linhao group of University of Electronic Science and technology of China has developed a tool for predicting protein lysine lactation sites DeepKla
▸▸▸▸
iMeta | Chinese Academy of Sciences Li Xiaofang et al. Dietary licorice promotes cadmium detoxification in mice and regulates intestinal flora metabolism
▸▸▸▸
iMeta | Ni Yan formation of Zhejiang University MetOrigin Trace the source of modern metabolites and analyze the integration of intestinal microbiome and metabolome
Journal Introduction

“iMeta” By Willie 、 Open access journal published by Enterobacteriaceae branch in cooperation with hundreds of Chinese scientists in this field , The editor in chief is Liu Shuangjiang, a researcher of the Institute of Microbiology of the Chinese Academy of Sciences, and Fu Jingyuan, a professor of Groningen University in the Netherlands . The purpose is to publish original research 、 Methods and reviews to promote metagenomics 、 Development of microbiome and bioinformatics . The goal is to publish 10%(IF > 15) High impact papers . Journal features include video contributions 、 Repeatable analysis 、 Picture polishing 、 Young editorial board members 、 front 3 Annual free publishing fee 、50 Social media publicity of 10000 users, etc .2022 year 2 It was officially launched in January !
Contact us
iMeta Home page :http://www.imeta.science
Press. :https://onlinelibrary.wiley.com/journal/2770596x
contribute :https://mc.manuscriptcentral.com/imeta
mailbox :[email protected]
WeChat official account
iMeta
editor
Slightly
边栏推荐
- 零代码高回报,如何用40套模板,能满足工作中95%的报表需求
- A way of writing SQL, update when matching, or insert
- 亚朵三顾 IPO
- Supersocket 1.6 creates a simple socket server with message length in the header
- DAY FIVE
- SuperSocket 1.6 创建一个简易的报文长度在头部的Socket服务器
- [boutique] Pinia Persistence Based on the plug-in Pinia plugin persist
- 达晨史上最大单笔投资,今天IPO了
- C语言输入/输出流和文件操作【二】
- 1000字精选 —— 接口测试基础
猜你喜欢
What is AVL tree?
Gradle knowledge generalization
Introduction to GPIO
![[unmanned aerial vehicle] multi unmanned cooperative task allocation program platform, including Matlab code](/img/4c/5d867437aac5faa299817e187602e1.png)
[unmanned aerial vehicle] multi unmanned cooperative task allocation program platform, including Matlab code
Asset security issues or constraints on the development of the encryption industry, risk control + compliance has become the key to breaking the platform

DAY FOUR
MATLIB从excel表中读取数据并画出函数图像
Pytest multi process / multi thread execution test case

Yaduo Sangu IPO
DAY ONE
随机推荐
DAY THREE
SQL的一种写法,匹配就更新,否则就是插入
Scholar doctor hahaha
PostgreSQL高可用之repmgr(1主2从+1witness)+Pgpool-II实现主从切换+读写分离
[CVPR 2022] semi supervised object detection: dense learning based semi supervised object detection
互动滑轨屏演示能为企业展厅带来什么
Design of short chain
ldap创建公司组织、人员
Wind chime card issuing network source code latest version - commercially available
快讯 l Huobi Ventures与Genesis公链深入接洽中
编译logisim
MySQL主从之多源复制(3主1从)搭建及同步测试
使用源码编译来安装PostgreSQL13.3数据库
matplotlib画柱状图并添加数值到图中
Typescript incremental compilation
There are only two TXT cells in the ArrayExpress database. Can you only download the sequencing run matrix from line to ENA?
快手的新生意,还得靠辛巴吆喝?
Tourism Management System Based on jsp+servlet+mysql framework [source code + database + report]
在docker中快速使用各个版本的PostgreSQL数据库
内网穿透zerotier 外网(手机、电脑等)访问内网设备(树莓派、NAS、电脑等)