当前位置:网站首页>Digital analog 1232
Digital analog 1232
2022-07-27 03:41:00 【Zoo is very true】
Adaptation race
subject 1
Random forests

Add a row BMI Parameters
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm']/100)**2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14,' Whether you get sick ', d)
print(df3.head(5))

import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
from sklearn import metrics
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm']/100)**2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14,' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# The first question is only to analyze metadata
df = df3[[' Gender ', ' Age ', ' height cm', ' weight kg', 'BMI',' Whether you get sick ']]
print(df.head(5))
# step3 model
dataset = pd.get_dummies(df, columns=[' Gender ', ' Whether you get sick ']) # Convert the classified variable into a heat independent variable
# print(dataset.head(5))
standardScaler = StandardScaler()
columns_to_scale = [' Age ', ' height cm', ' weight kg','BMI' ]
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head())
y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# print(y) # target The goal is
X = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# print(X) # Input characteristics
# Divide the training set and the test set
x_train, x_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=1)
# Create an instance of a random forest classifier
randomforest = RandomForestClassifier(random_state=42, n_estimators=100)
# Use the training set samples to train the classifier model
model = randomforest.fit(x_train, y_train)
# print(model.predict(x_train))
# print(y_train)
# print(type(model.predict(x_train)))
y_train = numpy.array(y_train)
xm = confusion_matrix(y_train.argmax(axis=1),model.predict(x_train).argmax(axis=1)) # Confusion matrix
print(' The training set confusion matrix is :\n',xm)
print(' Training set accuracy :', metrics.accuracy_score(model.predict(x_train), y_train))
y_test = numpy.array(y_test)
y_pred = model.predict(x_test)
cm = confusion_matrix(y_test.argmax(axis=1),y_pred.argmax(axis=1)) # Confusion matrix
print(' The confusion matrix of the test set is :\n',cm)
print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))
'''
The training set confusion matrix is :
[[1102 16]
[ 17 1102]]
Training set accuracy : 0.9852481001341081
The confusion matrix of the test set is :
[[261 221]
[239 239]]
Test set accuracy : 0.5166666666666667
'''SVM
Be careful SVM The category distribution of the dataset of cannot be different guo

import pandas as pd
import numpy as np
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn import svm
from sklearn import metrics
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1)
# print(df1) # [4923 rows x 13 columns] Read negative Too much leads to data imbalance
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm']/100)**2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14,' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# The first question is only to analyze metadata
df = df3[[' height cm',' Age ', ' weight kg', 'BMI',' Whether you get sick ']]
print(df.head(5))
# step3 model
# dataset = pd.get_dummies(df, columns=[' Gender ', ' Whether you get sick ']) # Convert the classified variable into a heat independent variable
dataset = df
# print(dataset.head(5))
standardScaler = StandardScaler()
columns_to_scale = [' height cm',' Age ', ' weight kg', 'BMI']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head())
y = dataset[[' Whether you get sick ']]
# print(y) # target The goal is
X = dataset.drop([' Whether you get sick '],axis=1)
# print(X) # Input characteristics
# Divide the training set and the test set
x_train, x_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=1)
# Create a SVM classifier
model = svm.SVC() # establish SVM classifier
model = model.fit(x_train, y_train) # Training with a training set
print(y_train)
prediction = model.predict(x_train) # Using test sets to predict
# p_svm = pd.DataFrame(prediction)
# p_svm.to_csv("train_svm.csv", index=False, sep=',')
# print(prediction)
print(' Training set accuracy :', metrics.accuracy_score(prediction, y_train))
prediction = model.predict(x_test) # Using test sets to predict
p_svm = pd.DataFrame(prediction)
p_svm.to_csv("test_svm.csv", index=False, sep=',')
print(' Test set accuracy :', metrics.accuracy_score(prediction, y_test))
neural network MLP

import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
from sklearn import metrics
from sklearn.neural_network import MLPClassifier
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm']/100)**2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14,' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# The first question is only to analyze metadata
df = df3[[' Gender ', ' Age ', ' height cm', ' weight kg', 'BMI',' Whether you get sick ']]
print(df.head(5))
# step3 model
dataset = pd.get_dummies(df, columns=[' Gender ', ' Whether you get sick ']) # Convert the classified variable into a heat independent variable
# print(dataset.head(5))
standardScaler = StandardScaler()
columns_to_scale = [' Age ', ' height cm', ' weight kg','BMI' ]
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head())
y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# print(y) # target The goal is
X = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# print(X) # Input characteristics
# Divide the training set and the test set
x_train, x_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=1)
mlp = MLPClassifier(solver='lbfgs',hidden_layer_sizes=(5,4),activation='logistic',max_iter=5000)
mlp.fit(x_train, y_train)
print(" Neural network method ")
y_t = mlp.predict(x_train)
print(' Training set accuracy :', metrics.accuracy_score(y_t, y_train))
y_pred = mlp.predict(x_test)
print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))
subject 2
Random forests
import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
ColNames_List = df3.columns.values.tolist()
# print('------------------------------------------------------')
# print(ColNames_List,type(ColNames_List))
# [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ',
# ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# The second question is
df = df3[[' Tidal volume (L) ',' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']]
# Label category
set(df[' Whether you get sick ']) #{0,1} negative -0, positive -1
print(df.shape)
# Count missing values
print(df.isna().sum())
print(df.describe())
'''
# Tidal volume has a large number of empty values
(6520, 10)
Tidal volume (L) 1974
Forced vital capacity (%) 0
Central airway parameters (%) 0
FEV1/FVC(%) 0
Maximum expiratory flow (%) 0
Peripheral airway parameters A(%) 0
Peripheral airway parameters B(%) 0
Peripheral airway parameters C(%) 0
Peripheral airway parameters D(%) 0
Whether you get sick 0
dtype: int64
Tidal volume (L) Forced vital capacity (%) ... Peripheral airway parameters D(%) Whether you get sick
count 4546.000000 6520.000000 ... 6520.000000 6520.000000
mean 1.388097 98.463413 ... 83.906838 0.244939
std 0.558965 11.734124 ... 33.676596 0.430084
min 0.160000 68.711656 ... 0.000000 0.000000
25% 0.990000 89.880350 ... 61.896197 0.000000
50% 1.310000 97.291561 ... 79.493590 0.000000
75% 1.730000 105.834854 ... 101.187574 0.000000
max 4.070000 188.603989 ... 679.464286 1.000000
[8 rows x 10 columns]
Process finished with exit code 0
'''
# Fill in missing values with means
mean_val =df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
print(' After filling ','\n',df.head(5))
print(df.isna().sum())
'''
1636 0.650000 106.319703 110.917031 ... 84.887460 62.626263 0
901 1.020000 108.368201 111.500000 ... 91.600000 61.160714 0
478 0.810000 100.000000 99.130435 ... 111.003861 93.534483 0
6145 1.388097 127.960526 124.809160 ... 69.047619 33.913043 1
4401 1.060000 83.266932 91.866029 ... 80.276134 73.660714 0
[5 rows x 10 columns]
Tidal volume (L) 0
Forced vital capacity (%) 0
Central airway parameters (%) 0
FEV1/FVC(%) 0
Maximum expiratory flow (%) 0
Peripheral airway parameters A(%) 0
Peripheral airway parameters B(%) 0
Peripheral airway parameters C(%) 0
Peripheral airway parameters D(%) 0
Whether you get sick 0
dtype: int64
'''
# step3 model
dataset = pd.get_dummies(df, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
standardScaler = StandardScaler()
columns_to_scale = [' Tidal volume (L) ',' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# print(X) # Input characteristics
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
feat_labels = df.columns[0:9] # The name of the feature
# print(feat_labels)
forest = RandomForestClassifier(n_estimators=10000, random_state=0, n_jobs=-1,max_depth=3)
forest.fit(x_train, y_train)
score = forest.score(x_test, y_test) # score=0.98148
importances = forest.feature_importances_ # The random forest model considers the importance of training characteristics
indices = np.argsort(importances)[::-1] # Subscript sort
for f in range(x_train.shape[1]): # x_train.shape[1]
print("%2d) %-*s %f" % \
(f + 1, 30, feat_labels[indices[f]], importances[indices[f]]))
'''
1) FEV1/FVC(%) 0.573146
2) Peripheral airway parameters A(%) 0.172901
3) Peripheral airway parameters B(%) 0.079313
4) Central airway parameters (%) 0.068985
5) Tidal volume (L) 0.037866
6) Forced vital capacity (%) 0.033652
7) Maximum expiratory flow (%) 0.022079
8) Peripheral airway parameters D(%) 0.006881
9) Peripheral airway parameters C(%) 0.005177
Choose the front 5 The effect of one may be better
'''
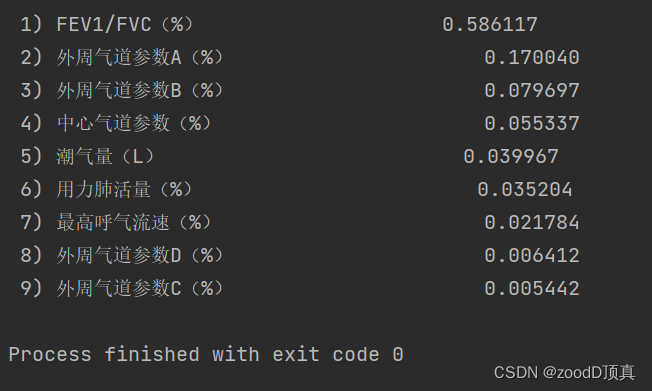
Let's first look at the prediction effect of the model without making a choice

import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
from sklearn import metrics
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
ColNames_List = df3.columns.values.tolist()
# print('------------------------------------------------------')
# print(ColNames_List,type(ColNames_List))
# [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ',
# ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# The second question is
df = df3[[' Tidal volume (L) ',' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']]
# Label category
set(df[' Whether you get sick ']) #{0,1} negative -0, positive -1
print(df.shape)
# Count missing values
print(df.isna().sum())
print(df.describe())
'''
# Tidal volume has a large number of empty values
(6520, 10)
Tidal volume (L) 1974
Forced vital capacity (%) 0
Central airway parameters (%) 0
FEV1/FVC(%) 0
Maximum expiratory flow (%) 0
Peripheral airway parameters A(%) 0
Peripheral airway parameters B(%) 0
Peripheral airway parameters C(%) 0
Peripheral airway parameters D(%) 0
Whether you get sick 0
dtype: int64
Tidal volume (L) Forced vital capacity (%) ... Peripheral airway parameters D(%) Whether you get sick
count 4546.000000 6520.000000 ... 6520.000000 6520.000000
mean 1.388097 98.463413 ... 83.906838 0.244939
std 0.558965 11.734124 ... 33.676596 0.430084
min 0.160000 68.711656 ... 0.000000 0.000000
25% 0.990000 89.880350 ... 61.896197 0.000000
50% 1.310000 97.291561 ... 79.493590 0.000000
75% 1.730000 105.834854 ... 101.187574 0.000000
max 4.070000 188.603989 ... 679.464286 1.000000
[8 rows x 10 columns]
Process finished with exit code 0
'''
# Fill in missing values with means
mean_val =df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
print(' After filling ','\n',df.head(5))
print(df.isna().sum())
'''
1636 0.650000 106.319703 110.917031 ... 84.887460 62.626263 0
901 1.020000 108.368201 111.500000 ... 91.600000 61.160714 0
478 0.810000 100.000000 99.130435 ... 111.003861 93.534483 0
6145 1.388097 127.960526 124.809160 ... 69.047619 33.913043 1
4401 1.060000 83.266932 91.866029 ... 80.276134 73.660714 0
[5 rows x 10 columns]
Tidal volume (L) 0
Forced vital capacity (%) 0
Central airway parameters (%) 0
FEV1/FVC(%) 0
Maximum expiratory flow (%) 0
Peripheral airway parameters A(%) 0
Peripheral airway parameters B(%) 0
Peripheral airway parameters C(%) 0
Peripheral airway parameters D(%) 0
Whether you get sick 0
dtype: int64
'''
# step3 model
dataset = pd.get_dummies(df, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
standardScaler = StandardScaler()
columns_to_scale = [' Tidal volume (L) ',' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# print(X) # Input characteristics
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
# Create an instance of a random forest classifier
randomforest = RandomForestClassifier(random_state=42, n_estimators=500)
# Use the training set samples to train the classifier model
model = randomforest.fit(x_train, y_train)
# print(model.predict(x_train))
# print(y_train)
# print(type(model.predict(x_train)))
y_train = numpy.array(y_train)
xm = confusion_matrix(y_train.argmax(axis=1),model.predict(x_train).argmax(axis=1)) # Confusion matrix
print(' The training set confusion matrix is :\n',xm)
print(' Training set accuracy :', metrics.accuracy_score(model.predict(x_train), y_train))
y_test = numpy.array(y_test)
y_pred = model.predict(x_test)
cm = confusion_matrix(y_test.argmax(axis=1),y_pred.argmax(axis=1)) # Confusion matrix
print(' The confusion matrix of the test set is :\n',cm)
print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))Make feature selection , Choose the former 7 individual

import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
from sklearn import metrics
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
ColNames_List = df3.columns.values.tolist()
# print('------------------------------------------------------')
# print(ColNames_List,type(ColNames_List))
# [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ',
# ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# The second question is
df = df3[[' Tidal volume (L) ',' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']]
# Label category
set(df[' Whether you get sick ']) #{0,1} negative -0, positive -1
print(df.shape)
# Count missing values
print(df.isna().sum())
print(df.describe())
'''
# Tidal volume has a large number of empty values
(6520, 10)
Tidal volume (L) 1974
Forced vital capacity (%) 0
Central airway parameters (%) 0
FEV1/FVC(%) 0
Maximum expiratory flow (%) 0
Peripheral airway parameters A(%) 0
Peripheral airway parameters B(%) 0
Peripheral airway parameters C(%) 0
Peripheral airway parameters D(%) 0
Whether you get sick 0
dtype: int64
Tidal volume (L) Forced vital capacity (%) ... Peripheral airway parameters D(%) Whether you get sick
count 4546.000000 6520.000000 ... 6520.000000 6520.000000
mean 1.388097 98.463413 ... 83.906838 0.244939
std 0.558965 11.734124 ... 33.676596 0.430084
min 0.160000 68.711656 ... 0.000000 0.000000
25% 0.990000 89.880350 ... 61.896197 0.000000
50% 1.310000 97.291561 ... 79.493590 0.000000
75% 1.730000 105.834854 ... 101.187574 0.000000
max 4.070000 188.603989 ... 679.464286 1.000000
[8 rows x 10 columns]
Process finished with exit code 0
'''
# Fill in missing values with means
mean_val =df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
print(' After filling ','\n',df.head(5))
print(df.isna().sum())
'''
1636 0.650000 106.319703 110.917031 ... 84.887460 62.626263 0
901 1.020000 108.368201 111.500000 ... 91.600000 61.160714 0
478 0.810000 100.000000 99.130435 ... 111.003861 93.534483 0
6145 1.388097 127.960526 124.809160 ... 69.047619 33.913043 1
4401 1.060000 83.266932 91.866029 ... 80.276134 73.660714 0
[5 rows x 10 columns]
Tidal volume (L) 0
Forced vital capacity (%) 0
Central airway parameters (%) 0
FEV1/FVC(%) 0
Maximum expiratory flow (%) 0
Peripheral airway parameters A(%) 0
Peripheral airway parameters B(%) 0
Peripheral airway parameters C(%) 0
Peripheral airway parameters D(%) 0
Whether you get sick 0
dtype: int64
'''
# step3 model
# Make feature selection
df = df[['FEV1/FVC(%)',' Peripheral airway parameters A(%)',' Peripheral airway parameters B(%)',' Central airway parameters (%)',' Tidal volume (L) ',' Forced vital capacity (%)',' Maximum expiratory flow (%)',' Whether you get sick ']]
print(df.head(5))
'''
FEV1/FVC(%) Peripheral airway parameters A(%) Peripheral airway parameters B(%) ... Forced vital capacity (%) Maximum expiratory flow (%) Whether you get sick
1491 88.235294 56.373938 91.962617 ... 82.156134 89.898990 0
80 79.794521 45.555556 91.546763 ... 97.333333 92.721519 0
296 82.951654 84.552846 94.329897 ... 85.249458 116.040956 0
2187 87.401575 66.954023 67.620751 ... 82.200647 76.863354 1
1071 86.764706 68.318966 99.506579 ... 97.142857 101.744186 0
'''
dataset = pd.get_dummies(df, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
standardScaler = StandardScaler()
columns_to_scale = ['FEV1/FVC(%)',' Peripheral airway parameters A(%)',' Peripheral airway parameters B(%)',' Central airway parameters (%)',' Tidal volume (L) ',' Forced vital capacity (%)',' Maximum expiratory flow (%)']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# print(X) # Input characteristics
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
# Create an instance of a random forest classifier
randomforest = RandomForestClassifier(random_state=42, n_estimators=500)
# Use the training set samples to train the classifier model
model = randomforest.fit(x_train, y_train)
# print(model.predict(x_train))
# print(y_train)
# print(type(model.predict(x_train)))
y_train = numpy.array(y_train)
xm = confusion_matrix(y_train.argmax(axis=1),model.predict(x_train).argmax(axis=1)) # Confusion matrix
print(" After feature selection ")
print(' The training set confusion matrix is :\n',xm)
print(' Training set accuracy :', metrics.accuracy_score(model.predict(x_train), y_train))
y_test = numpy.array(y_test)
y_pred = model.predict(x_test)
cm = confusion_matrix(y_test.argmax(axis=1),y_pred.argmax(axis=1)) # Confusion matrix
print(' The confusion matrix of the test set is :\n',cm)
print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))
'''
The training set confusion matrix is :
[[1119 0]
[ 0 1118]]
Training set accuracy : 1.0
The confusion matrix of the test set is :
[[372 109]
[156 323]]
Test set accuracy : 0.7166666666666667
'''svm

import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn import svm
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
ColNames_List = df3.columns.values.tolist()
# print('------------------------------------------------------')
# print(ColNames_List,type(ColNames_List))
# [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ',
# ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# The second question is
df = df3[[' Tidal volume (L) ',' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']]
# Label category
set(df[' Whether you get sick ']) #{0,1} negative -0, positive -1
print(df.shape)
# Count missing values
print(df.isna().sum())
print(df.describe())
'''
# Tidal volume has a large number of empty values
(6520, 10)
Tidal volume (L) 1974
Forced vital capacity (%) 0
Central airway parameters (%) 0
FEV1/FVC(%) 0
Maximum expiratory flow (%) 0
Peripheral airway parameters A(%) 0
Peripheral airway parameters B(%) 0
Peripheral airway parameters C(%) 0
Peripheral airway parameters D(%) 0
Whether you get sick 0
dtype: int64
Tidal volume (L) Forced vital capacity (%) ... Peripheral airway parameters D(%) Whether you get sick
count 4546.000000 6520.000000 ... 6520.000000 6520.000000
mean 1.388097 98.463413 ... 83.906838 0.244939
std 0.558965 11.734124 ... 33.676596 0.430084
min 0.160000 68.711656 ... 0.000000 0.000000
25% 0.990000 89.880350 ... 61.896197 0.000000
50% 1.310000 97.291561 ... 79.493590 0.000000
75% 1.730000 105.834854 ... 101.187574 0.000000
max 4.070000 188.603989 ... 679.464286 1.000000
[8 rows x 10 columns]
Process finished with exit code 0
'''
# Fill in missing values with means
mean_val =df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
print(' After filling ','\n',df.head(5))
print(df.isna().sum())
'''
1636 0.650000 106.319703 110.917031 ... 84.887460 62.626263 0
901 1.020000 108.368201 111.500000 ... 91.600000 61.160714 0
478 0.810000 100.000000 99.130435 ... 111.003861 93.534483 0
6145 1.388097 127.960526 124.809160 ... 69.047619 33.913043 1
4401 1.060000 83.266932 91.866029 ... 80.276134 73.660714 0
[5 rows x 10 columns]
Tidal volume (L) 0
Forced vital capacity (%) 0
Central airway parameters (%) 0
FEV1/FVC(%) 0
Maximum expiratory flow (%) 0
Peripheral airway parameters A(%) 0
Peripheral airway parameters B(%) 0
Peripheral airway parameters C(%) 0
Peripheral airway parameters D(%) 0
Whether you get sick 0
dtype: int64
'''
# step3 model
# Make feature selection
df = df[['FEV1/FVC(%)',' Peripheral airway parameters A(%)',' Peripheral airway parameters B(%)',' Central airway parameters (%)',' Tidal volume (L) ',' Forced vital capacity (%)',' Maximum expiratory flow (%)',' Whether you get sick ']]
print(df.head(5))
'''
FEV1/FVC(%) Peripheral airway parameters A(%) Peripheral airway parameters B(%) ... Forced vital capacity (%) Maximum expiratory flow (%) Whether you get sick
1491 88.235294 56.373938 91.962617 ... 82.156134 89.898990 0
80 79.794521 45.555556 91.546763 ... 97.333333 92.721519 0
296 82.951654 84.552846 94.329897 ... 85.249458 116.040956 0
2187 87.401575 66.954023 67.620751 ... 82.200647 76.863354 1
1071 86.764706 68.318966 99.506579 ... 97.142857 101.744186 0
'''
# dataset = pd.get_dummies(df, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
dataset = df
standardScaler = StandardScaler()
columns_to_scale = ['FEV1/FVC(%)',' Peripheral airway parameters A(%)',' Peripheral airway parameters B(%)',' Central airway parameters (%)',' Tidal volume (L) ',' Forced vital capacity (%)',' Maximum expiratory flow (%)']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick ']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick '],axis=1)
# print(X) # Input characteristics
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
model = svm.SVC() # establish SVM classifier
model = model.fit(x_train, y_train) # Training with a training set
print(y_train)
prediction = model.predict(x_train) # Using test sets to predict
# p_svm = pd.DataFrame(prediction)
# p_svm.to_csv("train_svm.csv", index=False, sep=',')
# print(prediction)
print(' Training set accuracy :', metrics.accuracy_score(prediction, y_train))
prediction = model.predict(x_test) # Using test sets to predict
p_svm = pd.DataFrame(prediction)
p_svm.to_csv("test_svm.csv", index=False, sep=',')
print(' Test set accuracy :', metrics.accuracy_score(prediction, y_test))
MLP
import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
from sklearn import metrics
from sklearn.neural_network import MLPClassifier
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
ColNames_List = df3.columns.values.tolist()
# print('------------------------------------------------------')
# print(ColNames_List,type(ColNames_List))
# [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ',
# ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# The second question is
df = df3[[' Tidal volume (L) ',' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']]
# Label category
set(df[' Whether you get sick ']) #{0,1} negative -0, positive -1
print(df.shape)
# Count missing values
print(df.isna().sum())
print(df.describe())
'''
# Tidal volume has a large number of empty values
(6520, 10)
Tidal volume (L) 1974
Forced vital capacity (%) 0
Central airway parameters (%) 0
FEV1/FVC(%) 0
Maximum expiratory flow (%) 0
Peripheral airway parameters A(%) 0
Peripheral airway parameters B(%) 0
Peripheral airway parameters C(%) 0
Peripheral airway parameters D(%) 0
Whether you get sick 0
dtype: int64
Tidal volume (L) Forced vital capacity (%) ... Peripheral airway parameters D(%) Whether you get sick
count 4546.000000 6520.000000 ... 6520.000000 6520.000000
mean 1.388097 98.463413 ... 83.906838 0.244939
std 0.558965 11.734124 ... 33.676596 0.430084
min 0.160000 68.711656 ... 0.000000 0.000000
25% 0.990000 89.880350 ... 61.896197 0.000000
50% 1.310000 97.291561 ... 79.493590 0.000000
75% 1.730000 105.834854 ... 101.187574 0.000000
max 4.070000 188.603989 ... 679.464286 1.000000
[8 rows x 10 columns]
Process finished with exit code 0
'''
# Fill in missing values with means
mean_val =df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
print(' After filling ','\n',df.head(5))
print(df.isna().sum())
'''
1636 0.650000 106.319703 110.917031 ... 84.887460 62.626263 0
901 1.020000 108.368201 111.500000 ... 91.600000 61.160714 0
478 0.810000 100.000000 99.130435 ... 111.003861 93.534483 0
6145 1.388097 127.960526 124.809160 ... 69.047619 33.913043 1
4401 1.060000 83.266932 91.866029 ... 80.276134 73.660714 0
[5 rows x 10 columns]
Tidal volume (L) 0
Forced vital capacity (%) 0
Central airway parameters (%) 0
FEV1/FVC(%) 0
Maximum expiratory flow (%) 0
Peripheral airway parameters A(%) 0
Peripheral airway parameters B(%) 0
Peripheral airway parameters C(%) 0
Peripheral airway parameters D(%) 0
Whether you get sick 0
dtype: int64
'''
# step3 model
# Make feature selection
df = df[['FEV1/FVC(%)',' Peripheral airway parameters A(%)',' Peripheral airway parameters B(%)',' Central airway parameters (%)',' Tidal volume (L) ',' Forced vital capacity (%)',' Maximum expiratory flow (%)',' Whether you get sick ']]
print(df.head(5))
'''
FEV1/FVC(%) Peripheral airway parameters A(%) Peripheral airway parameters B(%) ... Forced vital capacity (%) Maximum expiratory flow (%) Whether you get sick
1491 88.235294 56.373938 91.962617 ... 82.156134 89.898990 0
80 79.794521 45.555556 91.546763 ... 97.333333 92.721519 0
296 82.951654 84.552846 94.329897 ... 85.249458 116.040956 0
2187 87.401575 66.954023 67.620751 ... 82.200647 76.863354 1
1071 86.764706 68.318966 99.506579 ... 97.142857 101.744186 0
'''
dataset = pd.get_dummies(df, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
standardScaler = StandardScaler()
columns_to_scale = ['FEV1/FVC(%)',' Peripheral airway parameters A(%)',' Peripheral airway parameters B(%)',' Central airway parameters (%)',' Tidal volume (L) ',' Forced vital capacity (%)',' Maximum expiratory flow (%)']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# print(X) # Input characteristics
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
mlp = MLPClassifier(solver='lbfgs',hidden_layer_sizes=(5,4),activation='logistic',max_iter=5000)
mlp.fit(x_train, y_train)
print(" Neural network method ")
y_t = mlp.predict(x_train)
print(' Training set accuracy :', metrics.accuracy_score(y_t, y_train))
y_pred = mlp.predict(x_test)
print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))
Topic three
Random forests

import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
from sklearn import metrics
from sklearn import svm
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
ColNames_List = df3.columns.values.tolist()
# print('------------------------------------------------------')
# print(ColNames_List,type(ColNames_List))
# [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ',
# ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# Third question Metadata selection age 、 weight 、 height 、BMI、 Check whether the parameter is selected 7 individual
# drop_features = [' Gender ',' Peripheral airway parameters C(%)',' Peripheral airway parameters C(%)']
# df = df3.drop(drop_features, axis=1)
df = df3
# Fill in missing values with means
mean_val =df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
print(df.head(10))
print(df.columns)
'''
Index([' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)',
'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)',
' Peripheral airway parameters D(%)', 'BMI', ' Whether you get sick '],
dtype='object')
'''
# step3 model
dataset = pd.get_dummies(df, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
standardScaler = StandardScaler()
columns_to_scale = [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)',
'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)',
' Peripheral airway parameters D(%)', 'BMI']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# print(X) # Input characteristics
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
# Create an instance of a random forest classifier
randomforest = RandomForestClassifier(random_state=42, n_estimators=500)
# Use the training set samples to train the classifier model
model = randomforest.fit(x_train, y_train)
# print(model.predict(x_train))
# print(y_train)
# print(type(model.predict(x_train)))
y_train = numpy.array(y_train)
xm = confusion_matrix(y_train.argmax(axis=1),model.predict(x_train).argmax(axis=1)) # Confusion matrix
# print(" After feature selection ")
print(' The training set confusion matrix is :\n',xm)
print(' Training set accuracy :', metrics.accuracy_score(model.predict(x_train), y_train))
y_test = numpy.array(y_test)
y_pred = model.predict(x_test)
cm = confusion_matrix(y_test.argmax(axis=1),y_pred.argmax(axis=1)) # Confusion matrix
print(' The confusion matrix of the test set is :\n',cm)
print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))
'''
[5 rows x 16 columns]
The training set confusion matrix is :
[[1130 0]
[ 0 1107]]
Training set accuracy : 1.0
The confusion matrix of the test set is :
[[380 90]
[154 336]]
Test set accuracy : 0.74375
'''SVM

import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
from sklearn import metrics
from sklearn import svm
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
ColNames_List = df3.columns.values.tolist()
# print('------------------------------------------------------')
# print(ColNames_List,type(ColNames_List))
# [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ',
# ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# Third question Metadata selection age 、 weight 、 height 、BMI、 Check whether the parameter is selected 7 individual
# drop_features = [' Gender ',' Peripheral airway parameters C(%)',' Peripheral airway parameters C(%)']
# df = df3.drop(drop_features, axis=1)
df = df3
# Fill in missing values with means
mean_val =df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
print(df.head(10))
print(df.columns)
'''
Index([' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)',
'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)',
' Peripheral airway parameters D(%)', 'BMI', ' Whether you get sick '],
dtype='object')
'''
# step3 model
dataset = df
standardScaler = StandardScaler()
columns_to_scale = [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)',
'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)',
' Peripheral airway parameters D(%)', 'BMI']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick ']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick '],axis=1)
# print(X) # Input characteristics
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
model = svm.SVC() # establish SVM classifier
model = model.fit(x_train, y_train) # Training with a training set
print(y_train)
prediction = model.predict(x_train) # Using test sets to predict
# p_svm = pd.DataFrame(prediction)
# p_svm.to_csv("train_svm.csv", index=False, sep=',')
# print(prediction)
print(' Training set accuracy :', metrics.accuracy_score(prediction, y_train))
prediction = model.predict(x_test) # Using test sets to predict
p_svm = pd.DataFrame(prediction)
p_svm.to_csv("test_svm.csv", index=False, sep=',')
print(' Test set accuracy :', metrics.accuracy_score(prediction, y_test))
After feature selection
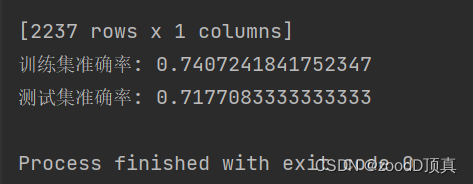
import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
from sklearn import metrics
from sklearn import svm
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
ColNames_List = df3.columns.values.tolist()
# print('------------------------------------------------------')
# print(ColNames_List,type(ColNames_List))
# [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ',
# ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# Third question Metadata selection age 、 weight 、 height 、BMI、 Check whether the parameter is selected 7 individual
drop_features = [' Gender ',' Peripheral airway parameters C(%)',' Peripheral airway parameters C(%)']
df = df3.drop(drop_features, axis=1)
# Fill in missing values with means
mean_val =df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
print(df.head(10))
print(df.columns)
'''
Index([' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)',
' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters D(%)', 'BMI', ' Whether you get sick '],
dtype='object')
'''
# step3 model
dataset = df
standardScaler = StandardScaler()
columns_to_scale = [' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)',
' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters D(%)', 'BMI']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick ']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick '],axis=1)
# print(X) # Input characteristics
#
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
model = svm.SVC() # establish SVM classifier
model = model.fit(x_train, y_train) # Training with a training set
print(y_train)
prediction = model.predict(x_train) # Using test sets to predict
# p_svm = pd.DataFrame(prediction)
# p_svm.to_csv("train_svm.csv", index=False, sep=',')
# print(prediction)
print(' Training set accuracy :', metrics.accuracy_score(prediction, y_train))
prediction = model.predict(x_test) # Using test sets to predict
p_svm = pd.DataFrame(prediction)
p_svm.to_csv("test_svm.csv", index=False, sep=',')
print(' Test set accuracy :', metrics.accuracy_score(prediction, y_test))neural network
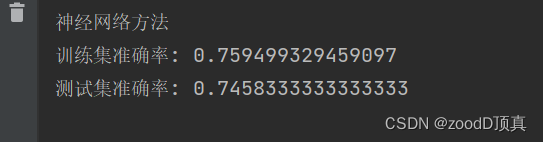
import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.neural_network import MLPClassifier
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
ColNames_List = df3.columns.values.tolist()
# print('------------------------------------------------------')
# print(ColNames_List,type(ColNames_List))
# [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ',
# ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# Third question Metadata selection age 、 weight 、 height 、BMI、 Check whether the parameter is selected 7 individual
drop_features = [' Gender ',' Peripheral airway parameters C(%)',' Peripheral airway parameters C(%)']
df = df3.drop(drop_features, axis=1)
# Fill in missing values with means
mean_val =df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
print(df.head(10))
print(df.columns)
'''
Index([' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)',
' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters D(%)', 'BMI', ' Whether you get sick '],
dtype='object')
'''
# step3 model
dataset = pd.get_dummies(df, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
standardScaler = StandardScaler() # Data standardization
columns_to_scale = [' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)',
' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters D(%)', 'BMI']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# print(X) # Input characteristics
#
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
mlp = MLPClassifier(solver='lbfgs',hidden_layer_sizes=(5,4),activation='logistic',max_iter=5000)
mlp.fit(x_train, y_train)
print(" Neural network method ")
y_t = mlp.predict(x_train)
print(' Training set accuracy :', metrics.accuracy_score(y_t, y_train))
y_pred = mlp.predict(x_test)
print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))K Crossover verification

import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
from sklearn import metrics
from sklearn import svm
from sklearn.model_selection import cross_val_score
from sklearn.neural_network import MLPClassifier
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
# df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
ColNames_List = df3.columns.values.tolist()
# print('------------------------------------------------------')
# print(ColNames_List,type(ColNames_List))
# [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ',
# ' Forced vital capacity (%)', ' Central airway parameters (%)', 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)', ' Whether you get sick ']
# print(df3)
'''
Gender Age height cm weight kg ... Peripheral airway parameters B(%) Peripheral airway parameters C(%) Peripheral airway parameters D(%) Whether you get sick
0 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
1 male 55 151.0 49.0 ... 59.870550 101.844262 83.653846 0
2 male 30 181.0 69.0 ... 64.519906 103.146067 107.100592 0
3 male 25 179.0 75.0 ... 62.237762 104.092072 92.814371 0
4 male 23 171.0 59.0 ... 54.024390 74.943567 48.076923 0
... .. .. ... ... ... ... ... ... ...
6515 Woman 36 167.0 60.0 ... 96.875000 102.512563 100.000000 1
6516 male 28 183.0 68.0 ... 102.870264 63.942308 82.269504 1
6517 Woman 36 160.0 55.0 ... 64.957265 58.203125 54.585153 1
6518 Woman 60 159.0 74.0 ... 100.957854 72.000000 48.529412 1
6519 Woman 63 156.0 46.0 ... 66.336634 92.553191 87.368421 1
[6520 rows x 14 columns]
'''
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# print(df3.head(5))
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
# print(df3)
# Third question Metadata selection age 、 weight 、 height 、BMI、 Check whether the parameter is selected 7 individual
# drop_features = [' Gender ',' Peripheral airway parameters C(%)',' Peripheral airway parameters C(%)']
# df = df3.drop(drop_features, axis=1)
df = df3
# Fill in missing values with means
mean_val =df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
print(df.head(10))
print(df.columns)
'''
Index([' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)',
'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)',
' Peripheral airway parameters D(%)', 'BMI', ' Whether you get sick '],
dtype='object')
'''
# step3 model
dataset = pd.get_dummies(df, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
standardScaler = StandardScaler()
columns_to_scale = [' Gender ', ' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)',
'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', ' Peripheral airway parameters C(%)',
' Peripheral airway parameters D(%)', 'BMI']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# print(X) # Input characteristics
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
# Split the training set from the test set , Note that all cross validation is done on the training set , Only the last part of the test set will be used
# Create a random forest instance
rf = RandomForestClassifier(random_state=42, n_estimators=500)
rf_mse = cross_val_score(estimator = rf , X = x_train, y = y_train, scoring = 'r2', cv = 5, verbose = 1, n_jobs=6)
print(' In the random forest model ,R2 The average is %.4f, The standard deviation is %.4f' %(rf_mse.mean(), rf_mse.std()))
mlp = MLPClassifier(solver='lbfgs',hidden_layer_sizes=(5,4),activation='logistic',max_iter=5000)
mlp_mse = cross_val_score(estimator = mlp, X = x_train, y = y_train, scoring = 'r2', cv = 5, verbose = 1, n_jobs=6)
print('MLP In the model ,R2 The average is %.4f, The standard deviation is %.4f' %(mlp_mse.mean(), mlp_mse.std()))
# The average is the highest ( Low deviation ), The standard deviation is the smallest ( Low variance ) Is a good model
# In the random forest model ,R2 The average is -0.2770, The standard deviation is 0.0515
# MLP In the model ,R2 The average is -0.3461, The standard deviation is 0.0922
# Therefore, the model is selected as Random forests Random forest parameter optimization

# Parameter optimization Yes n_estimators Parameter optimization
randomforest = RandomForestClassifier(random_state=42)
param_test1 = {"n_estimators": range(1, 101, 10)}
gsearch1 = GridSearchCV(estimator=RandomForestClassifier(), param_grid=param_test1,
scoring='roc_auc', cv=10)
gsearch1.fit(x_train, y_train)
print(gsearch1.best_score_)
print(gsearch1.best_params_)
print("best accuracy:%f" % gsearch1.best_score_)
'''
0.7900814280926616
{'n_estimators': 81}
best accuracy:0.790081
''' 
# Parameter optimization Maximum characteristic number max_features, Other parameters are set to constant , And n_estimators by 81
randomforest = RandomForestClassifier(random_state=42)
param_test2 = {"max_features":range(1,11,1)}
gsearch1 = GridSearchCV(estimator=RandomForestClassifier(n_estimators=81,
random_state=10),
param_grid = param_test2,scoring='roc_auc',cv=10)
gsearch1.fit(x_train, y_train)
print(gsearch1.best_score_)
print(gsearch1.best_params_)
print("best accuracy:%f" % gsearch1.best_score_)
'''
0.7900814280926616
{'n_estimators': 81}
best accuracy:0.790081
0.7973612760484702
{'max_features': 9}
best accuracy:0.797361
'''Compare the optimized results
Nothing changed. , Draft

randomforest = RandomForestClassifier(random_state=42,n_estimators=81,max_features=9)
model = randomforest.fit(x_train, y_train)
# print(model.predict(x_train))
# print(y_train)
# print(type(model.predict(x_train)))
y_train = numpy.array(y_train)
xm = confusion_matrix(y_train.argmax(axis=1),model.predict(x_train).argmax(axis=1)) # Confusion matrix
# print(" After feature selection ")
print(' The training set confusion matrix is :\n',xm)
print(' Training set accuracy :', metrics.accuracy_score(model.predict(x_train), y_train))
y_test = numpy.array(y_test)
y_pred = model.predict(x_test)
cm = confusion_matrix(y_test.argmax(axis=1),y_pred.argmax(axis=1)) # Confusion matrix
print(' The confusion matrix of the test set is :\n',cm)
print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))
'''
The training set confusion matrix is :
[[1108 0]
[ 0 1129]]
Training set accuracy : 1.0
The confusion matrix of the test set is :
[[393 99]
[147 321]]
Test set accuracy : 0.74375
'''
Sample imbalance problem

import numpy
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import confusion_matrix
from sklearn import metrics
from sklearn.metrics import recall_score # Recall rate
# step1 Data preprocessing
# Reading data
path = 'xiaochuan.xlsx'
xl = pd.ExcelFile(path)
# print(xl.sheet_names) # [' negative ', ' positive ']
df1 = xl.parse(' negative ', index_col=0)
# df1 = df1.loc[0:1600,:]
# print(df1) # [4923 rows x 13 columns] Read negative
df2 = xl.parse(' positive ', index_col=0)
# print(df2) # [1597 rows x 13 columns] Read positive data
# First, add a column respectively Are you sick negative 0 positive 1
df1.insert(loc=13, column=' Whether you get sick ', value=0) # Negative is 0
# print(df1) # 1 male 40 176.0 67.0 ... 64.661654 105.070423 76.190476 0
df2.insert(loc=13, column=' Whether you get sick ', value=1) # Positive is 1
# print(df2) # 1 male 20 174.0 77.0 ... 62.248521 67.021277 58.035714 1
# take df1 and df 2 Merge together
df3 = pd.concat([df1, df2], axis=0).reset_index(drop=True)
# increase BMI Parameters BMI= weight (kg)÷ height 2(m2).
df3['BMI'] = df3[' weight kg'] / ((df3[' height cm'] / 100) ** 2)
# print(3**2)
d = df3.pop(' Whether you get sick ')
df3.insert(14, ' Whether you get sick ', d)
# Handle the gender column
df3.replace(' male ', 1, inplace=True)
df3.replace(' Woman ', 0, inplace=True)
df3 = shuffle(df3)
drop_features = [' Gender ', ' Peripheral airway parameters C(%)', ' Peripheral airway parameters D(%)'] # Delete the unimportant features of previous relevance
df = df3.drop(drop_features, axis=1)
# Fill in missing values with means
mean_val = df[' Tidal volume (L) '].mean()
# print(mean_val) # 1.3880972283325999
df[' Tidal volume (L) '].fillna(mean_val, inplace=True)
# print(df.head(5))
# print(df.isna().sum())
dataset = df
x = dataset.iloc[:, :-1] # features
y = dataset.iloc[:, -1] # label
groupby_data_o = dataset.groupby([' Whether you get sick '])[' Whether you get sick '].count() # Label category classification count
# Analyze sample imbalance
print(groupby_data_o)
'''
Whether you get sick
0 4923
1 1597 Sample imbalance exists
Name: Whether you get sick , dtype: int64
'''
# Use oversampling
# Use SMOTE Methods sampling treatment was carried out
from imblearn.over_sampling import SMOTE # Oversampling Library SMOTE
model_smote = SMOTE()
x_smote_resampled, y_smote_resampled = model_smote.fit_resample(x, y) # The input data has been sampled
y_smote_resampled = pd.DataFrame(y_smote_resampled, columns=[' Whether you get sick '])
smote_resampled = pd.concat([x_smote_resampled, y_smote_resampled], axis=1) # Reassemble features and labels
group_data_smote = smote_resampled.groupby([' Whether you get sick '])[' Whether you get sick '].count() # Check the number of label categories
print(group_data_smote)
'''
Whether you get sick
0 4923
1 4923
Name: Whether you get sick , dtype: int64
'''
dataset = pd.get_dummies(smote_resampled, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
standardScaler = StandardScaler() # Standardization
columns_to_scale = [' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)',
'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', 'BMI']
dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
print(dataset.head(5))
y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# print(y) # target The goal is
x = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# print(X) # Input characteristics
# Random forests Regardless of sample classification imbalance
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
# Create an instance of a random forest classifier
randomforest = RandomForestClassifier(random_state=42,n_estimators=81,max_features=9)
# Use the training set samples to train the classifier model
model = randomforest.fit(x_train, y_train)
# print(model.predict(x_train))
# print(y_train)
# print(type(model.predict(x_train)))
y_train = numpy.array(y_train)
xm = confusion_matrix(y_train.argmax(axis=1),model.predict(x_train).argmax(axis=1)) # Confusion matrix
print(" After using oversampling ")
print(' The training set confusion matrix is :\n',xm)
print(' Training set accuracy :', metrics.accuracy_score(model.predict(x_train), y_train))
y_test = numpy.array(y_test)
y_pred = model.predict(x_test)
cm = confusion_matrix(y_test.argmax(axis=1),y_pred.argmax(axis=1)) # Confusion matrix
print(' The confusion matrix of the test set is :\n',cm)
print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))
'''
After using oversampling
The training set confusion matrix is :
[[3477 0]
[ 0 3415]]
Training set accuracy : 1.0
The confusion matrix of the test set is :
[[1130 316]
[ 226 1282]]
Test set accuracy : 0.8165199729180772
'''
# # Use RandomUnderSampler Conduct under sampling
# from imblearn.under_sampling import RandomUnderSampler # Under sampling processing library RandomUnderSampler
#
# model_RandomUnderSampler = RandomUnderSampler() # Instantiation
# x_RandomUnderSampler_resampled, y_RandomUnderSampler_resampled = model_RandomUnderSampler.fit_resample(x,y) # Input data for under sampling
# y_RandomUnderSampler_resampled = pd.DataFrame(y_RandomUnderSampler_resampled, columns=[' Whether you get sick '])
#
# RandomUnderSampler_resampled = pd.concat([x_RandomUnderSampler_resampled, y_RandomUnderSampler_resampled],
# axis=1) # Reassemble features and labels
# group_data_RandomUnderSampler = RandomUnderSampler_resampled.groupby([' Whether you get sick '])[' Whether you get sick '].count() # Check the number of label categories
#
# print(group_data_RandomUnderSampler)
#
# # RandomUnderSampler_resampled Sampled data
#
# dataset = pd.get_dummies(RandomUnderSampler_resampled, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
# standardScaler = StandardScaler() # Standardization
# columns_to_scale = [' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)',
# 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', 'BMI']
# dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
# print(dataset.head(5))
#
# y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# # print(y) # target The goal is
# x = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# # print(X) # Input characteristics
#
#
# # Random forests Regardless of sample classification imbalance
# x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
# # Create an instance of a random forest classifier
# randomforest = RandomForestClassifier(random_state=42,n_estimators=81,max_features=9)
# # Use the training set samples to train the classifier model
# model = randomforest.fit(x_train, y_train)
#
# # print(model.predict(x_train))
# # print(y_train)
# # print(type(model.predict(x_train)))
# y_train = numpy.array(y_train)
# xm = confusion_matrix(y_train.argmax(axis=1),model.predict(x_train).argmax(axis=1)) # Confusion matrix
# print(" After using undersampling ")
# print(' The training set confusion matrix is :\n',xm)
# print(' Training set accuracy :', metrics.accuracy_score(model.predict(x_train), y_train))
#
# y_test = numpy.array(y_test)
#
# y_pred = model.predict(x_test)
# cm = confusion_matrix(y_test.argmax(axis=1),y_pred.argmax(axis=1)) # Confusion matrix
#
# print(' The confusion matrix of the test set is :\n',cm)
# print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))
#
# '''
# After using undersampling
# The training set confusion matrix is :
# [[1089 0]
# [ 0 1146]]
# Training set accuracy : 1.0
# The confusion matrix of the test set is :
# [[321 187]
# [145 306]]
# Test set accuracy : 0.6538060479666319
# '''
# dataset = pd.get_dummies(df, columns=[' Whether you get sick ']) # Convert the classified variable into a heat independent variable
# standardScaler = StandardScaler() # Standardization
# columns_to_scale = [' Age ', ' height cm', ' weight kg', ' Tidal volume (L) ', ' Forced vital capacity (%)', ' Central airway parameters (%)',
# 'FEV1/FVC(%)', ' Maximum expiratory flow (%)', ' Peripheral airway parameters A(%)', ' Peripheral airway parameters B(%)', 'BMI']
# dataset[columns_to_scale] = standardScaler.fit_transform(dataset[columns_to_scale])
# print(dataset.head(5))
#
# y = dataset[[' Whether you get sick _0',' Whether you get sick _1']]
# # print(y) # target The goal is
# x = dataset.drop([' Whether you get sick _0',' Whether you get sick _1'],axis=1)
# # print(X) # Input characteristics
#
#
# # Random forests Regardless of sample classification imbalance
# x_train, x_test, y_train, y_test = train_test_split(x, y, test_size = 0.3, random_state = 0)
# # Create an instance of a random forest classifier
# randomforest = RandomForestClassifier(random_state=42,n_estimators=81,max_features=9)
# # Use the training set samples to train the classifier model
# model = randomforest.fit(x_train, y_train)
#
# # print(model.predict(x_train))
# # print(y_train)
# # print(type(model.predict(x_train)))
# y_train = numpy.array(y_train)
# xm = confusion_matrix(y_train.argmax(axis=1),model.predict(x_train).argmax(axis=1)) # Confusion matrix
# # print(" After feature selection ")
# print(' The training set confusion matrix is :\n',xm)
# print(' Training set accuracy :', metrics.accuracy_score(model.predict(x_train), y_train))
#
# y_test = numpy.array(y_test)
#
# y_pred = model.predict(x_test)
# cm = confusion_matrix(y_test.argmax(axis=1),y_pred.argmax(axis=1)) # Confusion matrix
#
# print(' The confusion matrix of the test set is :\n',cm)
# print(' Test set accuracy :', metrics.accuracy_score(y_pred, y_test))
# '''
# The training set confusion matrix is :
# [[3459 0]
# [ 0 1105]]
# Training set accuracy : 1.0
# The confusion matrix of the test set is :
# [[1362 102]
# [ 380 112]]
# Test set accuracy : 0.7535787321063395
# '''
plt.imshow(cm, cmap=plt.cm.Blues)
indices = range(len(cm))
plt.xticks(indices, [0,1])
plt.yticks(indices, [0,1])
plt.colorbar()
plt.xlabel('guess')
plt.ylabel('fact')
for first_index in range(len(cm)):
for second_index in range(len(cm[first_index])):
plt.text(first_index, second_index, cm[second_index][first_index])
plt.title(' Optimize the confusion matrix of random forest test set ')
plt.show()边栏推荐
- 如何进行 360 评估
- Design method and test method of APP interface use case
- spark学习笔记(四)——sparkcore核心编程-RDD
- 【常用搜索问题】111
- How to uniquely identify a user SQL in Youxuan database cluster
- Spark: calculate the average value of the same key in different partitions (entry level - simple implementation)
- 快速排序及优化
- Deployment of ruoyi's environment and operation of the system
- How to optimize MySQL
- Data analysis and disassembly method of banyan tree in Bairong
猜你喜欢

Spark Learning Notes (IV) -- spark core programming RDD

Practical application of digital twins: smart city project construction solution

The application and significance of digital twins are the main role and conceptual value of electric power.
It's too strong. An annotation handles the data desensitization returned by the interface

Customer cases | pay attention to the elderly user experience, and the transformation of bank app to adapt to aging should avoid falsehood and be practical

The function and application of lpci-252 universal PCI interface can card
Message rejected MQ

spark:地区广告点击量排行统计(小案例)

索引最佳实践
若依框架代码生成详解
随机推荐
app端接口用例设计方法和测试方法
复盘:图像有哪些基本属性?关于图像的知识你知道哪些?图像的参数有哪些
spark学习笔记(六)——sparkcore核心编程-RDD行动算子
Typescript TS basic knowledge interface, generics
Unity game, the simplest solution of privacy agreement! Just 3 lines of code! (Reprinted)
客户案例 | 关注老年用户体验,银行APP适老化改造要避虚就实
Mysql database related operations
Redis source code learning (33), command execution process
架构基本概念和架构本质
DNS record type and explanation of related terms
mysql如何优化
数字孪生应用及意义对电力的主要作用,概念价值。
redis秒杀案例,跟着b站尚硅谷老师学习
客户端发送一条sql如何与服务器交互
Redis spike case, learn from Shang Silicon Valley teacher in station B
2022牛客多校第二场的J -- 三分做法
Reading notes of Kazuo Inamori's advice to young people
【正则】判断, 手机号,身份证号
shell awk
Sqlserver select * can you exclude a field
