当前位置:网站首页>GGPUBR: HOW TO ADD ADJUSTED P-VALUES TO A MULTI-PANEL GGPLOT
GGPUBR: HOW TO ADD ADJUSTED P-VALUES TO A MULTI-PANEL GGPLOT
2022-07-02 09:38:00 【小宇2022】
# Load required R packages
library(ggpubr)
library(rstatix)
# Data preparation
df <- tibble::tribble(
~sample_type, ~expression, ~cancer_type, ~gene,
"cancer", 25.8, "Lung", "Gene1",
"cancer", 25.5, "Liver", "Gene1",
"cancer", 22.4, "Liver", "Gene1",
"cancer", 21.2, "Lung", "Gene1",
"cancer", 24.5, "Liver", "Gene1",
"cancer", 27.3, "Liver", "Gene1",
"cancer", 30.9, "Liver", "Gene1",
"cancer", 17.6, "Breast", "Gene1",
"cancer", 19.7, "Lung", "Gene1",
"cancer", 9.7, "Breast", "Gene1",
"cancer", 15.2, "Breast", "Gene2",
"cancer", 26.4, "Liver", "Gene2",
"cancer", 25.8, "Lung", "Gene2",
"cancer", 9.7, "Breast", "Gene2",
"cancer", 21.2, "Lung", "Gene2",
"cancer", 24.5, "Liver", "Gene2",
"cancer", 14.5, "Breast", "Gene2",
"cancer", 19.7, "Lung", "Gene2",
"cancer", 25.2, "Lung", "Gene2",
"normal", 43.5, "Lung", "Gene1",
"normal", 76.5, "Liver", "Gene1",
"normal", 21.9, "Breast", "Gene1",
"normal", 69.9, "Liver", "Gene1",
"normal", 101.7, "Liver", "Gene1",
"normal", 80.1, "Liver", "Gene1",
"normal", 19.2, "Breast", "Gene1",
"normal", 49.5, "Lung", "Gene1",
"normal", 34.5, "Breast", "Gene1",
"normal", 51.9, "Lung", "Gene1",
"normal", 67.5, "Lung", "Gene2",
"normal", 30, "Breast", "Gene2",
"normal", 76.5, "Liver", "Gene2",
"normal", 88.5, "Liver", "Gene2",
"normal", 69.9, "Liver", "Gene2",
"normal", 49.5, "Lung", "Gene2",
"normal", 80.1, "Liver", "Gene2",
"normal", 79.2, "Liver", "Gene2",
"normal", 12.6, "Breast", "Gene2",
"normal", 97.5, "Liver", "Gene2",
"normal", 64.5, "Liver", "Gene2"
)
# Summary statistics
df %>%
group_by(gene, cancer_type, sample_type) %>%
get_summary_stats(expression, type = "common")
# Statistical test
# group the data by cancer type and gene
# Compare expression values of normal and cancer samples
stat.test <- df %>%
group_by(cancer_type, gene) %>%
t_test(expression ~ sample_type) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance()
stat.test
# Create boxplot
bxp <- ggboxplot(
df, x = "sample_type", y = "expression",
facet.by = c("gene", "cancer_type")
) +
rotate_x_text(angle = 60)
# Add adjusted p-values
stat.test <- stat.test %>% add_xy_position(x = "sample_type")
bxp + stat_pvalue_manual(stat.test, label = "p.adj")

# Load required R packages
library(ggpubr)
library(rstatix)
# Data preparation
df <- tibble::tribble(
~sample_type, ~expression, ~cancer_type, ~gene,
"cancer", 25.8, "Lung", "Gene1",
"cancer", 25.5, "Liver", "Gene1",
"cancer", 22.4, "Liver", "Gene1",
"cancer", 21.2, "Lung", "Gene1",
"cancer", 24.5, "Liver", "Gene1",
"cancer", 27.3, "Liver", "Gene1",
"cancer", 30.9, "Liver", "Gene1",
"cancer", 17.6, "Breast", "Gene1",
"cancer", 19.7, "Lung", "Gene1",
"cancer", 9.7, "Breast", "Gene1",
"cancer", 15.2, "Breast", "Gene2",
"cancer", 26.4, "Liver", "Gene2",
"cancer", 25.8, "Lung", "Gene2",
"cancer", 9.7, "Breast", "Gene2",
"cancer", 21.2, "Lung", "Gene2",
"cancer", 24.5, "Liver", "Gene2",
"cancer", 14.5, "Breast", "Gene2",
"cancer", 19.7, "Lung", "Gene2",
"cancer", 25.2, "Lung", "Gene2",
"normal", 43.5, "Lung", "Gene1",
"normal", 76.5, "Liver", "Gene1",
"normal", 21.9, "Breast", "Gene1",
"normal", 69.9, "Liver", "Gene1",
"normal", 101.7, "Liver", "Gene1",
"normal", 80.1, "Liver", "Gene1",
"normal", 19.2, "Breast", "Gene1",
"normal", 49.5, "Lung", "Gene1",
"normal", 34.5, "Breast", "Gene1",
"normal", 51.9, "Lung", "Gene1",
"normal", 67.5, "Lung", "Gene2",
"normal", 30, "Breast", "Gene2",
"normal", 76.5, "Liver", "Gene2",
"normal", 88.5, "Liver", "Gene2",
"normal", 69.9, "Liver", "Gene2",
"normal", 49.5, "Lung", "Gene2",
"normal", 80.1, "Liver", "Gene2",
"normal", 79.2, "Liver", "Gene2",
"normal", 12.6, "Breast", "Gene2",
"normal", 97.5, "Liver", "Gene2",
"normal", 64.5, "Liver", "Gene2"
)
# Summary statistics
df %>%
group_by(gene, cancer_type, sample_type) %>%
get_summary_stats(expression, type = "common")
# Statistical test
# group the data by cancer type and gene
# Compare expression values of normal and cancer samples
stat.test <- df %>%
group_by(cancer_type, gene) %>%
t_test(expression ~ sample_type) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance()
stat.test
# Create boxplot
bxp <- ggboxplot(
df, x = "sample_type", y = "expression",
facet.by = c("gene", "cancer_type")
) +
rotate_x_text(angle = 60)
# Add adjusted p-values
stat.test <- stat.test %>% add_xy_position(x = "sample_type")
bxp + stat_pvalue_manual(stat.test, label = "p.adj.signif")

# Load required R packages
library(ggpubr)
library(rstatix)
# Data preparation
df <- tibble::tribble(
~sample_type, ~expression, ~cancer_type, ~gene,
"cancer", 25.8, "Lung", "Gene1",
"cancer", 25.5, "Liver", "Gene1",
"cancer", 22.4, "Liver", "Gene1",
"cancer", 21.2, "Lung", "Gene1",
"cancer", 24.5, "Liver", "Gene1",
"cancer", 27.3, "Liver", "Gene1",
"cancer", 30.9, "Liver", "Gene1",
"cancer", 17.6, "Breast", "Gene1",
"cancer", 19.7, "Lung", "Gene1",
"cancer", 9.7, "Breast", "Gene1",
"cancer", 15.2, "Breast", "Gene2",
"cancer", 26.4, "Liver", "Gene2",
"cancer", 25.8, "Lung", "Gene2",
"cancer", 9.7, "Breast", "Gene2",
"cancer", 21.2, "Lung", "Gene2",
"cancer", 24.5, "Liver", "Gene2",
"cancer", 14.5, "Breast", "Gene2",
"cancer", 19.7, "Lung", "Gene2",
"cancer", 25.2, "Lung", "Gene2",
"normal", 43.5, "Lung", "Gene1",
"normal", 76.5, "Liver", "Gene1",
"normal", 21.9, "Breast", "Gene1",
"normal", 69.9, "Liver", "Gene1",
"normal", 101.7, "Liver", "Gene1",
"normal", 80.1, "Liver", "Gene1",
"normal", 19.2, "Breast", "Gene1",
"normal", 49.5, "Lung", "Gene1",
"normal", 34.5, "Breast", "Gene1",
"normal", 51.9, "Lung", "Gene1",
"normal", 67.5, "Lung", "Gene2",
"normal", 30, "Breast", "Gene2",
"normal", 76.5, "Liver", "Gene2",
"normal", 88.5, "Liver", "Gene2",
"normal", 69.9, "Liver", "Gene2",
"normal", 49.5, "Lung", "Gene2",
"normal", 80.1, "Liver", "Gene2",
"normal", 79.2, "Liver", "Gene2",
"normal", 12.6, "Breast", "Gene2",
"normal", 97.5, "Liver", "Gene2",
"normal", 64.5, "Liver", "Gene2"
)
# Summary statistics
df %>%
group_by(gene, cancer_type, sample_type) %>%
get_summary_stats(expression, type = "common")
# Statistical test
# group the data by cancer type and gene
# Compare expression values of normal and cancer samples
stat.test <- df %>%
group_by(cancer_type, gene) %>%
t_test(expression ~ sample_type) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance()
stat.test
# Create boxplot
bxp <- ggboxplot(
df, x = "sample_type", y = "expression",
facet.by = c("gene", "cancer_type")
) +
rotate_x_text(angle = 60)
# Add adjusted p-values
stat.test <- stat.test %>% add_xy_position(x = "sample_type")
# Hide ns and change the bracket tip length
bxp + stat_pvalue_manual(
stat.test, label = "p.adj.signif",
hide.ns = TRUE, tip.length = 0
)

# Load required R packages
library(ggpubr)
library(rstatix)
# Data preparation
df <- tibble::tribble(
~sample_type, ~expression, ~cancer_type, ~gene,
"cancer", 25.8, "Lung", "Gene1",
"cancer", 25.5, "Liver", "Gene1",
"cancer", 22.4, "Liver", "Gene1",
"cancer", 21.2, "Lung", "Gene1",
"cancer", 24.5, "Liver", "Gene1",
"cancer", 27.3, "Liver", "Gene1",
"cancer", 30.9, "Liver", "Gene1",
"cancer", 17.6, "Breast", "Gene1",
"cancer", 19.7, "Lung", "Gene1",
"cancer", 9.7, "Breast", "Gene1",
"cancer", 15.2, "Breast", "Gene2",
"cancer", 26.4, "Liver", "Gene2",
"cancer", 25.8, "Lung", "Gene2",
"cancer", 9.7, "Breast", "Gene2",
"cancer", 21.2, "Lung", "Gene2",
"cancer", 24.5, "Liver", "Gene2",
"cancer", 14.5, "Breast", "Gene2",
"cancer", 19.7, "Lung", "Gene2",
"cancer", 25.2, "Lung", "Gene2",
"normal", 43.5, "Lung", "Gene1",
"normal", 76.5, "Liver", "Gene1",
"normal", 21.9, "Breast", "Gene1",
"normal", 69.9, "Liver", "Gene1",
"normal", 101.7, "Liver", "Gene1",
"normal", 80.1, "Liver", "Gene1",
"normal", 19.2, "Breast", "Gene1",
"normal", 49.5, "Lung", "Gene1",
"normal", 34.5, "Breast", "Gene1",
"normal", 51.9, "Lung", "Gene1",
"normal", 67.5, "Lung", "Gene2",
"normal", 30, "Breast", "Gene2",
"normal", 76.5, "Liver", "Gene2",
"normal", 88.5, "Liver", "Gene2",
"normal", 69.9, "Liver", "Gene2",
"normal", 49.5, "Lung", "Gene2",
"normal", 80.1, "Liver", "Gene2",
"normal", 79.2, "Liver", "Gene2",
"normal", 12.6, "Breast", "Gene2",
"normal", 97.5, "Liver", "Gene2",
"normal", 64.5, "Liver", "Gene2"
)
# Summary statistics
df %>%
group_by(gene, cancer_type, sample_type) %>%
get_summary_stats(expression, type = "common")
# Statistical test
# group the data by cancer type and gene
# Compare expression values of normal and cancer samples
stat.test <- df %>%
group_by(cancer_type, gene) %>%
t_test(expression ~ sample_type) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance()
stat.test
# Create boxplot
bxp <- ggboxplot(
df, x = "sample_type", y = "expression",
facet.by = c("gene", "cancer_type")
) +
rotate_x_text(angle = 60)
# Add adjusted p-values
stat.test <- stat.test %>% add_xy_position(x = "sample_type")
# Show p-values and significance levels
bxp + stat_pvalue_manual(
stat.test, label = "{p.adj}{p.adj.signif}",
hide.ns = TRUE, tip.length = 0
)
边栏推荐
- Programmer growth Chapter 6: how to choose a company?
- SSRF
- 基于 Openzeppelin 的可升级合约解决方案的注意事项
- Array splitting (regular thinking
- Tdsql | difficult employment? Tencent cloud database micro authentication to help you
- mysql 基本语句
- SQLite modify column type
- Is the stock account given by qiniu business school safe? Can I open an account?
- [cloud native] 2.5 kubernetes core practice (Part 2)
- Webauthn - official development document
猜你喜欢

Solve the problem of data blank in the quick sliding page of the uniapp list
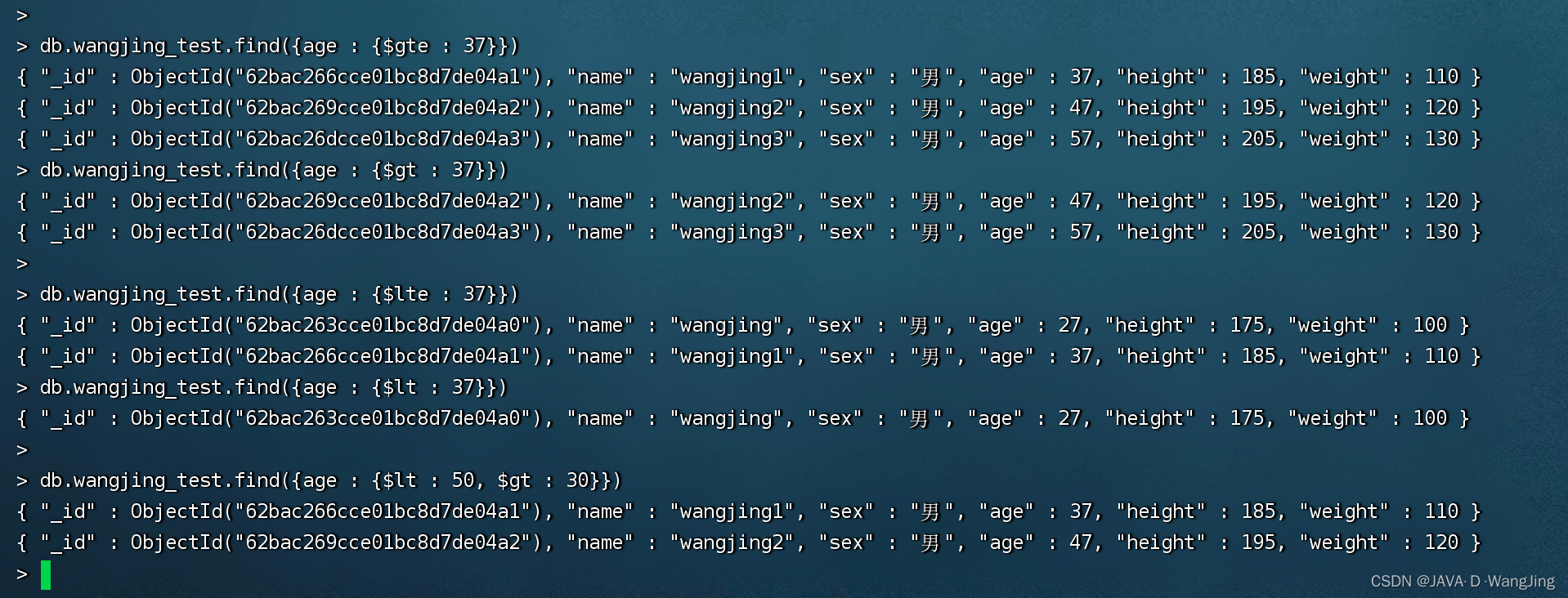
Mongodb learning and sorting (condition operator, $type operator, limit() method, skip() method and sort() method)

可昇級合約的原理-DelegateCall
I STM32 development environment, keil5/mdk5.14 installation tutorial (with download link)

CTF record
Skills of PLC recorder in quickly monitoring multiple PLC bits

C#多维数组的属性获取方法及操作注意

MTK full dump grab

ImportError: cannot import name ‘Digraph‘ from ‘graphviz‘
MySQL linked list data storage query sorting problem
随机推荐
tqdm的多行显示与单行显示
Tdsql | difficult employment? Tencent cloud database micro authentication to help you
spritejs
2022年4月17日五心红娘团队收获双份喜报
Is bond fund safe? Does the bond buying foundation lose principal?
Basic usage of MySQL in centos8
From the perspective of attack surface, see the practice of zero trust scheme of Xinchuang
sqlite 修改列类型
解决uniapp列表快速滑动页面数据空白问题
CTF record
揭露数据不一致的利器 —— 实时核对系统
Never forget, there will be echoes | hanging mirror sincerely invites you to participate in the opensca user award research
Regular and common formulas
On April 17, 2022, the five heart matchmaker team received double good news
What is the relationship between digital transformation of manufacturing industry and lean production
VS2019代码中包含中文内容导致的编译错误和打印输出乱码问题
Pit of the start attribute of enumrate
ctf 记录
Resources reads 2D texture and converts it to PNG format
ImportError: cannot import name ‘Digraph‘ from ‘graphviz‘
