当前位置:网站首页>Download the details and sequence of the original data access from the ENA database in EBI
Download the details and sequence of the original data access from the ENA database in EBI
2022-07-05 04:35:00 【Most afraid of being gentle】
List of articles
Biology students will probably use EBI database , And one of the ENA The database contains abundant sequence information , So how to get them ?
1. ENA The retrieval function of the database
ENA Database website :https://www.ebi.ac.uk/ena/browser/home
Before downloading information, the first thing is to retrieve information :
1. Simple search
Here's the picture , Search more directly sediment Information about , There will be a lot of relevant sequence information , search The search box can search all proteins ,RNA,DNA, Strain classification name ,accession Number, etc. .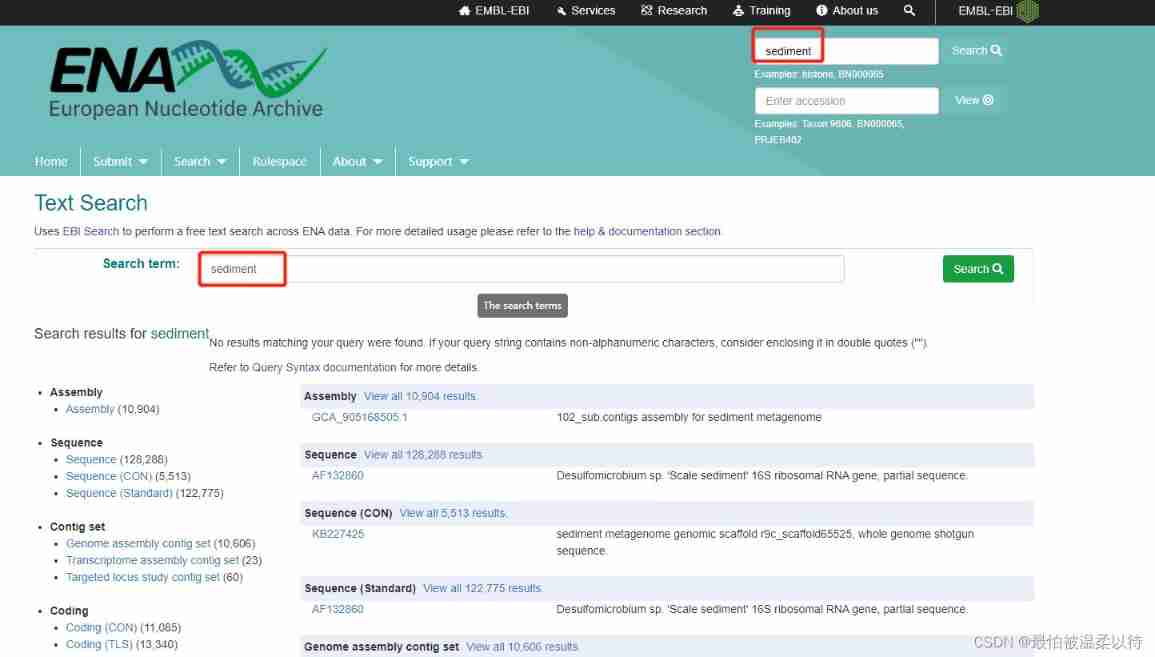
2. Complex search
Want all your more accurate , You need to use advanced search. stay search Drop down options in the column .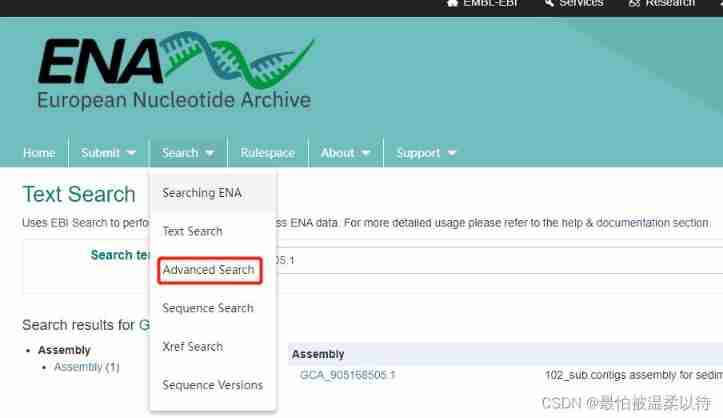
advance search It is divided into the following steps , Among them, as a condition screening is Data Type and Query Options .
(Query It can also be based on ENA Search syntax rules to write code search )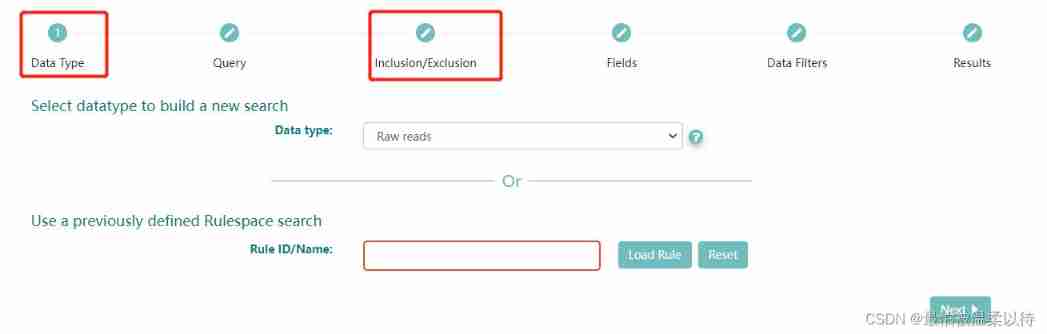
Data Type: Is the type of sequence you choose , Choose here Raw reads, The raw data of sequencing .Rule ID/Name Don't fill in .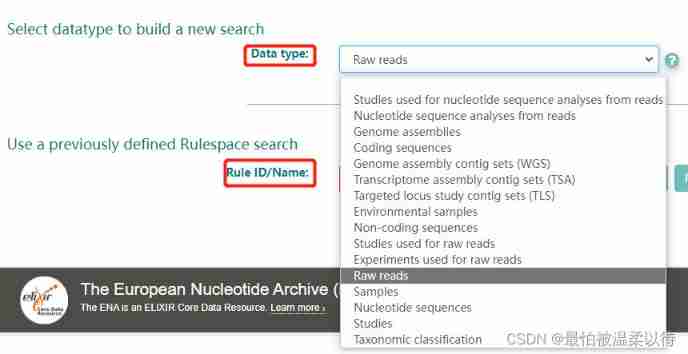
Query Options : Is the information condition of the sequence you want to filter . Select the corresponding condition in the opposite option box on the left .
for example : What I chose was libraray source and library strategy Namely metagenomic and wgs, That is to say, I want to screen the whole genome data of all macrogenomic tests . In addition to Query The corresponding... Will be generated in the box ENA Code of the database . Here, of course, you can also choose other conditions , Finally, click search Options will show results .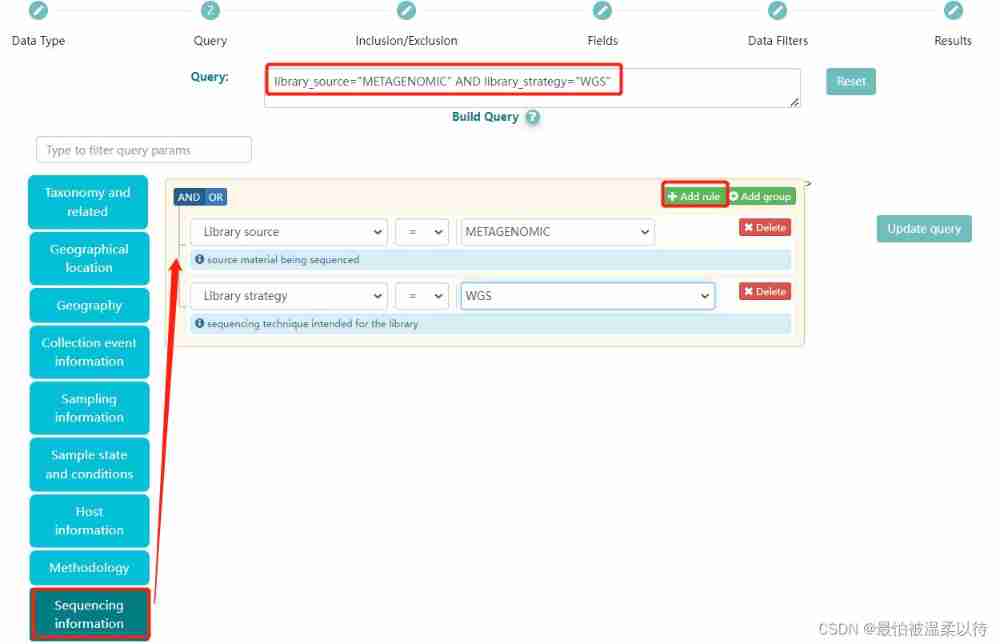
Download all results : My subsequent processing is based on downloading txt File to do . So click TSV Download to get the corresponding txx The file .
The result in the file is brief accession The number corresponds to title Information .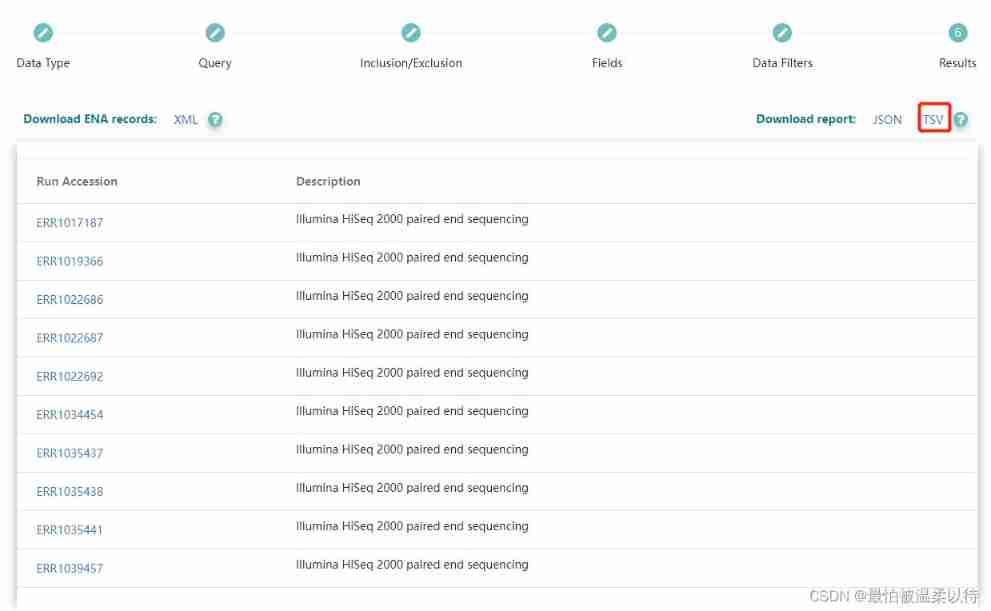
Results file :
2. ENA In the database accession Download the details of
I wrote this one myself python Script implementation . Attach the source code directly .
Because recursive and asynchronous downloading are used , Therefore, in the middle of the cycle, there will be error messages such as network request timeout , Don't worry about him , Just wait until the program is over . The program only needs to modify the file name in the last step .
# You can download the package you need
import aiohttp
import aiofiles
import asyncio
import requests
import pandas as pd
import time
import csv
""" 1. Based on ENA Database download txt file , Crawling accession Details of 2. The generated results are multiple output file , Then merge """
def read_file(file_name):
with open(file_name, 'r', encoding='utf-8')as f:
f = f.readlines()
accession_list = []
for i in f[1::]:
accession = i.strip().split('\t')[0].strip('"')
accession_list.append(accession)
return accession_list
async def get_biosample_info(accession_number, session, df):
headers = {
# "Referer": f'https://www.ebi.ac.uk/ena/browser/view/{accession_number}',
"User-Agent": "Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/96.0.4664.110 Safari/537.36",
}
params = {
"result": "read_run",
"accession": accession_number,
"offset": 0,
"limit": 1000,
"format": "json",
"fields": "study_accession,secondary_study_accession,sample_accession,secondary_sample_accession,experiment_accession,run_accession,submission_accession,tax_id,scientific_name,instrument_platform,instrument_model,library_name,nominal_length,library_layout,library_strategy,library_source,library_selection,read_count,base_count,center_name,first_public,last_updated,experiment_title,study_title,study_alias,experiment_alias,run_alias,fastq_bytes,fastq_md5,fastq_ftp,fastq_aspera,fastq_galaxy,submitted_bytes,submitted_md5,submitted_ftp,submitted_aspera,submitted_galaxy,submitted_format,sra_bytes,sra_md5,sra_ftp,sra_aspera,sra_galaxy,cram_index_ftp,cram_index_aspera,cram_index_galaxy,sample_alias,broker_name,sample_title,nominal_sdev,first_created"
}
url = r'https://www.ebi.ac.uk/ena/portal/api/filereport'
async with session.get(url, headers=headers, params=params) as resp:
if resp.content:
biosample_info_dic = await resp.json()
biosample_info_dic = biosample_info_dic[0]
print('now dealing with %s'%accession_number)
for k, v in biosample_info_dic.items():
df.loc[accession_number, k] = v
resp.close()
async def main(accession_list, num, input_file):
tasks = []
df = pd.DataFrame()
if accession_list:
# prevent ssl Report errors
timeout = aiohttp.ClientTimeout(total=600) # Set the timeout to 600 second
# force_close=True
connector = aiohttp.TCPConnector(limit=80, ssl=False) # Reduce the number of concurrent , no need ssl verification
async with aiohttp.ClientSession(connector=connector, timeout=timeout) as session:
for accession in accession_list:
task = asyncio.create_task(get_biosample_info(accession, session, df))
tasks.append(task)
await asyncio.wait(tasks)
print(df)
output_file = input_file.split('_')[0]+"_output"+str(num)+".xlsx"
df.to_excel(output_file)
# Find those that have not been downloaded accession Number
accessions = list(df.index)
not_download_accession = [i for i in accession_list if i not in accessions]
# Recursive download
await main(not_download_accession, int(num)+1, input_file)
if __name__ == '__main__':
try:
print(' Start the download ')
loop = asyncio.get_event_loop()
# Just modify the downloaded file name
loop.run_until_complete(main(read_file('results_read_run_tsv.txt'), 1, 'results_read_run_tsv.txt'))
loop.close()
print(" Download the end ")
except:
pass
The result is each accession Corresponding details .( You need to manually output Just merge the results of , Finally, I didn't bother to write a program to merge )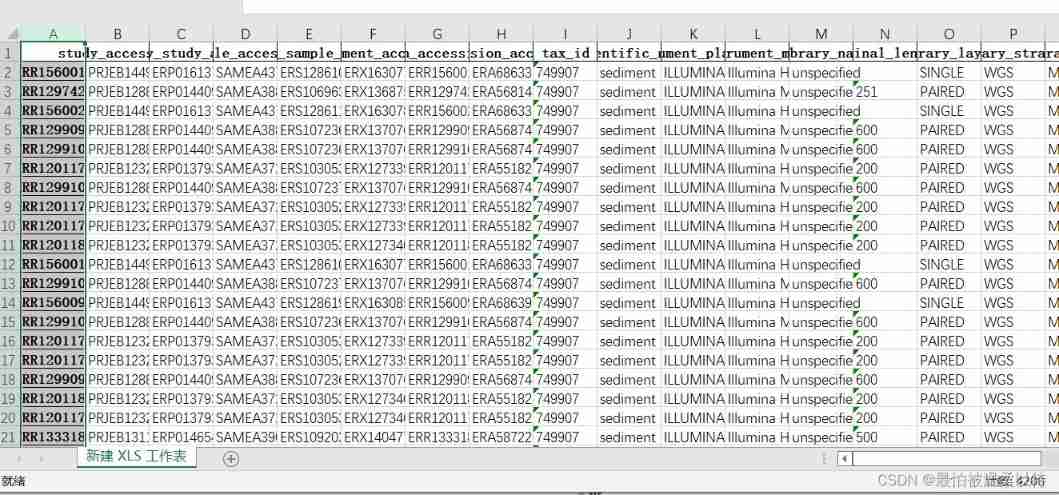
3. ENA In the database accession Download the sequence of
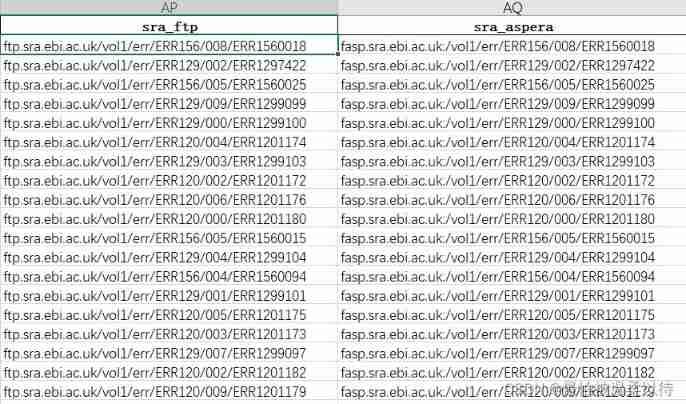
In the file sra_ftp and sra_aspera There will be a corresponding download address in , You can download in batches by integrating the website yourself .
边栏推荐
- Components in protective circuit
- [PCL self study: feature9] global aligned spatial distribution (GASD) descriptor (continuously updated)
- Burpsuite grabs app packets
- 49 pictures and 26 questions explain in detail what is WiFi?
- Raki's notes on reading paper: code and named entity recognition in stackoverflow
- Function (error prone)
- Introduction to RT thread kernel (4) -- clock management
- [finebi] the process of making custom maps using finebi
- [crampon programming] lintcode decoding Encyclopedia - 872 termination process
- Hypothesis testing -- learning notes of Chapter 8 of probability theory and mathematical statistics
猜你喜欢
![[moteur illusoire UE] il ne faut que six étapes pour réaliser le déploiement du flux de pixels ue5 et éviter les détours! (4.26 et 4.27 principes similaires)](/img/eb/a93630aff7545c6c3b71dcc9f5aa61.png)
[moteur illusoire UE] il ne faut que six étapes pour réaliser le déploiement du flux de pixels ue5 et éviter les détours! (4.26 et 4.27 principes similaires)

CUDA Programming atomic operation atomicadd reports error err:msb3721, return code 1

直播预告 | 容器服务 ACK 弹性预测最佳实践
windows下Redis-cluster集群搭建
Managed service network: application architecture evolution in the cloud native Era
函数(易错)

2022-2028 global and Chinese equipment as a Service Market Research Report
Sword finger offer 04 Search in two-dimensional array
User behavior collection platform
Sequence diagram of single sign on Certification Center
随机推荐
CSDN body auto generate directory
About the prompt loading after appscan is opened: guilogic, it keeps loading and gets stuck. My personal solution. (it may be the first solution available in the whole network at present)
【虚幻引擎UE】打包报错出现!FindPin错误的解决办法
包 类 包的作用域
Variable category (automatic, static, register, external)
【虛幻引擎UE】實現UE5像素流部署僅需六步操作少走彎路!(4.26和4.27原理類似)
How to remove installed elpa package
Function overloading
直播预告 | 容器服务 ACK 弹性预测最佳实践
American 5g open ran suffered another major setback, and its attempt to counter China's 5g technology has failed
Introduction to RT thread kernel (5) -- memory management
函数(易错)
[PCL self study: feature9] global aligned spatial distribution (GASD) descriptor (continuously updated)
Practice | mobile end practice
All in one 1413: determine base
取余操作是一个哈希函数
Decryption function calculates "task state and lifecycle management" of asynchronous task capability
Stage experience
Leetcode hot topic Hot 100 day 33: "subset"
Managed service network: application architecture evolution in the cloud native Era