当前位置:网站首页>Using tansformer to segment three-dimensional abdominal multiple organs -- actual battle of unetr
Using tansformer to segment three-dimensional abdominal multiple organs -- actual battle of unetr
2022-07-07 10:37:00 【Sister Tina】
Can't transformer No problem , This tutorial is out of the box .
Tina Sister is finally right transformer Let's go , I felt difficult before , Because I didn't learn it when I first learned sequence model . Then I have been rejecting learning transformer.
Nothing happened these two weeks , add MONAI There are ready-made tutorials , I plan to get through first and then , Learn the theory again . then , Successfully ran through the code , I learned theory for another week , It's nothing more than that , It's easy to get started .
Some students want to know transformer Words , You can finish this practical tutorial first , If you are interested , There will be a follow-up transformer Entry route .
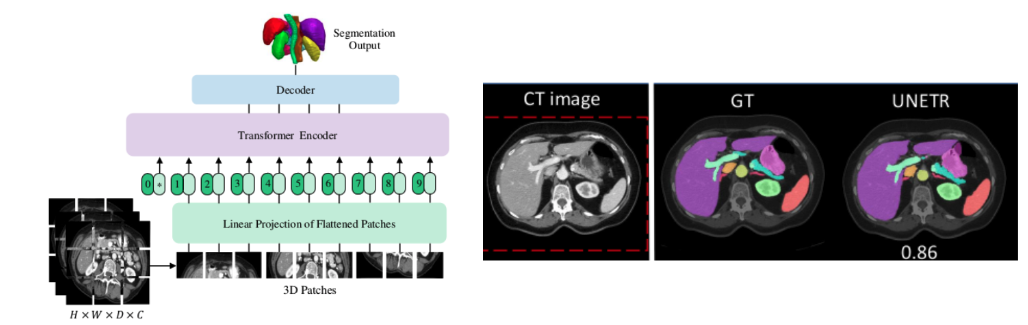
UNETR Introduce
Pure utilization Transformers As an encoder, it can learn the sequence representation of input quantity and effectively capture the global multi-scale information . At the same time, it also follows the successful of encoder and decoder “U type ” Network design .Transformers The encoder is directly connected to the decoder through hopping connections of different resolutions , To get the final segmentation result .
Using multiple organ segmentation BTCV Data sets 、 Medical split decathlon (MSD) The data set widely validates the proposed model in different imaging methods ( namely MR and CT) On the performance of volume brain tumor and spleen segmentation task , And the results always prove good performance .
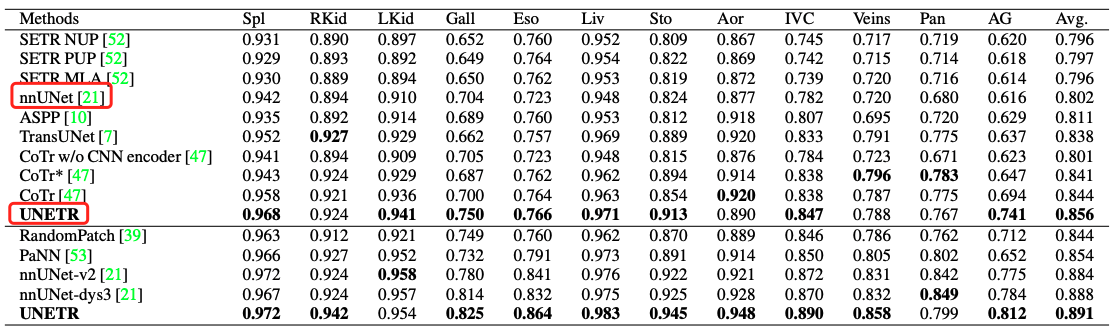
In the paper , The segmentation results of each data set are as follows
- BTCV Abdominal multiple organ segmentation results
- MSD On dataset : Spleen segmentation and brain tumor segmentation
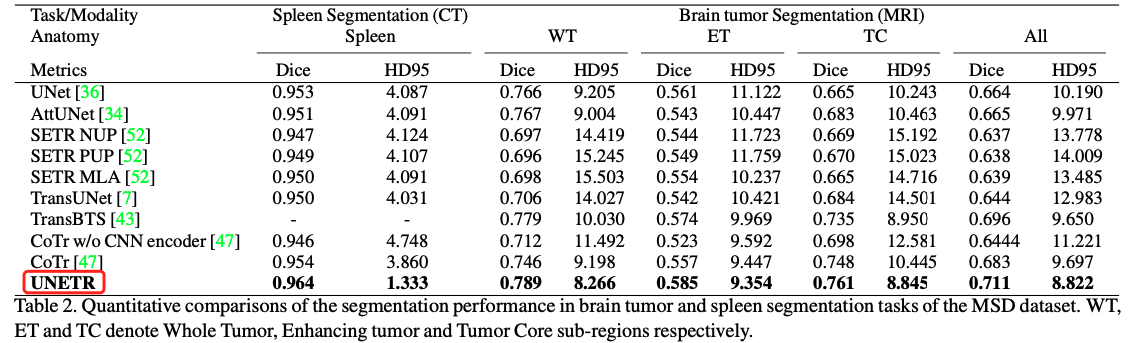
You can see ,UNETR Defeated on multiple abdominal organs nnUet.
UNETR Model structure of 
If I haven't learned Transformer, Just take this picture as Unet Look at , On the left is the down sampling , The size keeps shrinking , On the right is the upper sample , The size keeps expanding . In the middle is the jumping connection .
But the bottom sampling here is Transformer.
With a general understanding , Return to our actual combat
The actual combat stage
This tutorial code connects :MONAI UNETR tutorial
Download it , Watch my commentary while running , Better effect 🤭
It mainly includes the following parts :
- Conversion of dictionary format data .
- Data enhancement transformation : according to MONAI transform API Define a new transform.
- Load data from folder .
- cache IO And transformation to speed up training and verification .
- 3D UNETR Model 、DiceCE Loss function 、 Average of multiple organ segmentation tasks Dice Measure .
First , Download data , Data address :BTCV challenge round 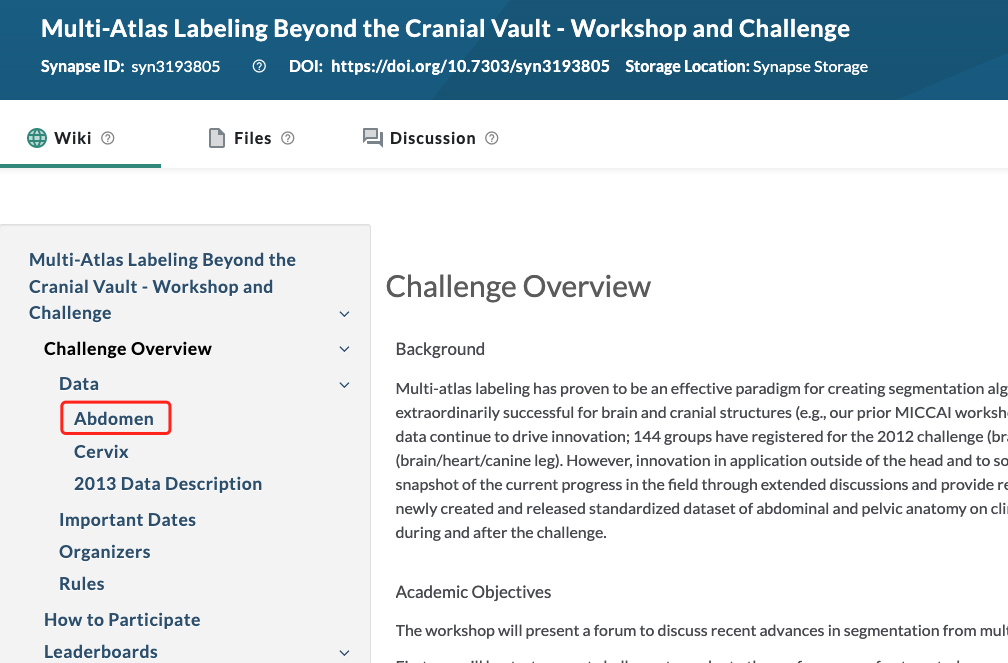
Be careful : Go through this link , It's hard to find data , I looked for it, too 2 Genius finds , I forgot exactly how to click in . It seems to be about to join the competition , To get to the place where the data is downloaded .
however , Never mind . Cannot find download connection , You can go to my network disk to download :
link : https://pan.baidu.com/s/1-0yMfZ4grBF5UYlRp1t_Rw Extraction code : ejfp
Understand the data
On the institutional review committee (IRB) Under the supervision of , Randomly selected from the combination of ongoing chemotherapy trials for colorectal cancer and retrospective abdominal hernia studies 50 Abdomen CT scanning . among ,30 For training ,20 For testing .
- Volume :512 x 512 x 85 - 512 x 512 x 198
- View :280 x 280 x 280 mm3 - 500 x 500 x 650 mm3
- Plane resolution :0.54 x 0.54 mm2 - 0.98 x 0.98 mm2
- z Axis resolution :2.5 mm To 5.0 mm
- target: A total of segmentation 13 Seed organ :1. spleen 2. Right kidney 3. Left kidney 4. Gallbladder 5. The esophagus 6. The liver 7. The stomach 8. The aorta 9. Inferior vena cava 10. Portal vein and splenic vein 11. pancreas 12 Right adrenal gland 13 Left adrenal gland .
- Training set 30 Data , It is divided into 24 Training + 6 validation
Environmental preparation
Make sure MONAI Version in 0.6 above , It's best to update to the latest version .
If your environment ok, This step can be omitted . Don't run
Load all necessary packages
import os
import shutil
import tempfile
import matplotlib.pyplot as plt
import numpy as np
from tqdm import tqdm
from monai.losses import DiceCELoss
from monai.inferers import sliding_window_inference
from monai.transforms import (
AsDiscrete,
AddChanneld,
Compose,
CropForegroundd,
LoadImaged,
Orientationd,
RandFlipd,
RandCropByPosNegLabeld,
RandShiftIntensityd,
ScaleIntensityRanged,
Spacingd,
RandRotate90d,
ToTensord,
)
from monai.config import print_config
from monai.metrics import DiceMetric
from monai.networks.nets import UNETR
from monai.data import (
DataLoader,
CacheDataset,
load_decathlon_datalist,
decollate_batch,
)
import torch
print_config()
This step , If you are prompted what package is missing , Just install what package
Set the model saving environment
root_dir = './checkpoints'
if not os.path.exists(root_dir):
os.makedirs(root_dir)
print(root_dir)
The original tutorial uses a temporary address , Here we change , Save the trained model
Set the training set and verification set transform
train_transforms = Compose(
[
LoadImaged(keys=["image", "label"]),
AddChanneld(keys=["image", "label"]),
Orientationd(keys=["image", "label"], axcodes="RAS"),
Spacingd(
keys=["image", "label"],
pixdim=(1.5, 1.5, 2.0),
mode=("bilinear", "nearest"),
),
ScaleIntensityRanged(
keys=["image"],
a_min=-175,
a_max=250,
b_min=0.0,
b_max=1.0,
clip=True,
),
CropForegroundd(keys=["image", "label"], source_key="image"),
RandCropByPosNegLabeld(
keys=["image", "label"],
label_key="label",
spatial_size=(96, 96, 96),
pos=1,
neg=1,
num_samples=4,
image_key="image",
image_threshold=0,
),
RandFlipd(
keys=["image", "label"],
spatial_axis=[0],
prob=0.10,
),
RandFlipd(
keys=["image", "label"],
spatial_axis=[1],
prob=0.10,
),
RandFlipd(
keys=["image", "label"],
spatial_axis=[2],
prob=0.10,
),
RandRotate90d(
keys=["image", "label"],
prob=0.10,
max_k=3,
),
RandShiftIntensityd(
keys=["image"],
offsets=0.10,
prob=0.50,
),
ToTensord(keys=["image", "label"]),
]
)
val_transforms = Compose(
[
LoadImaged(keys=["image", "label"]),
AddChanneld(keys=["image", "label"]),
Orientationd(keys=["image", "label"], axcodes="RAS"),
Spacingd(
keys=["image", "label"],
pixdim=(1.5, 1.5, 2.0),
mode=("bilinear", "nearest"),
),
ScaleIntensityRanged(
keys=["image"], a_min=-175, a_max=250, b_min=0.0, b_max=1.0, clip=True
),
CropForegroundd(keys=["image", "label"], source_key="image"),
ToTensord(keys=["image", "label"]),
]
)
You can see ,transformer This part of is the same as CNN There is no difference between . How to deal with data and how to deal with .
Download the data as required
I have already introduced how to download data , Here, make sure that the data meets the format required by the code .
Unzip the downloaded data , On the project ./data Next 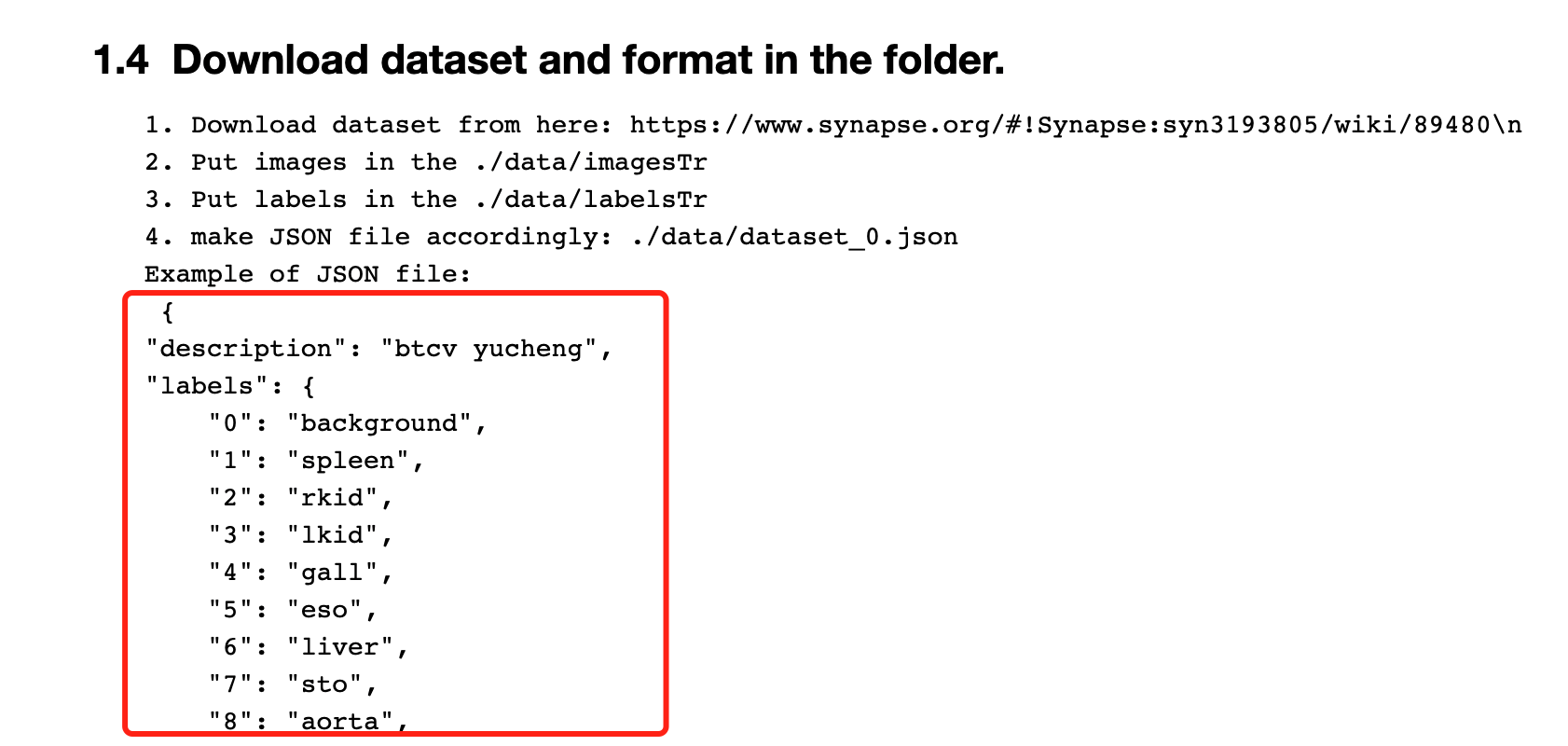
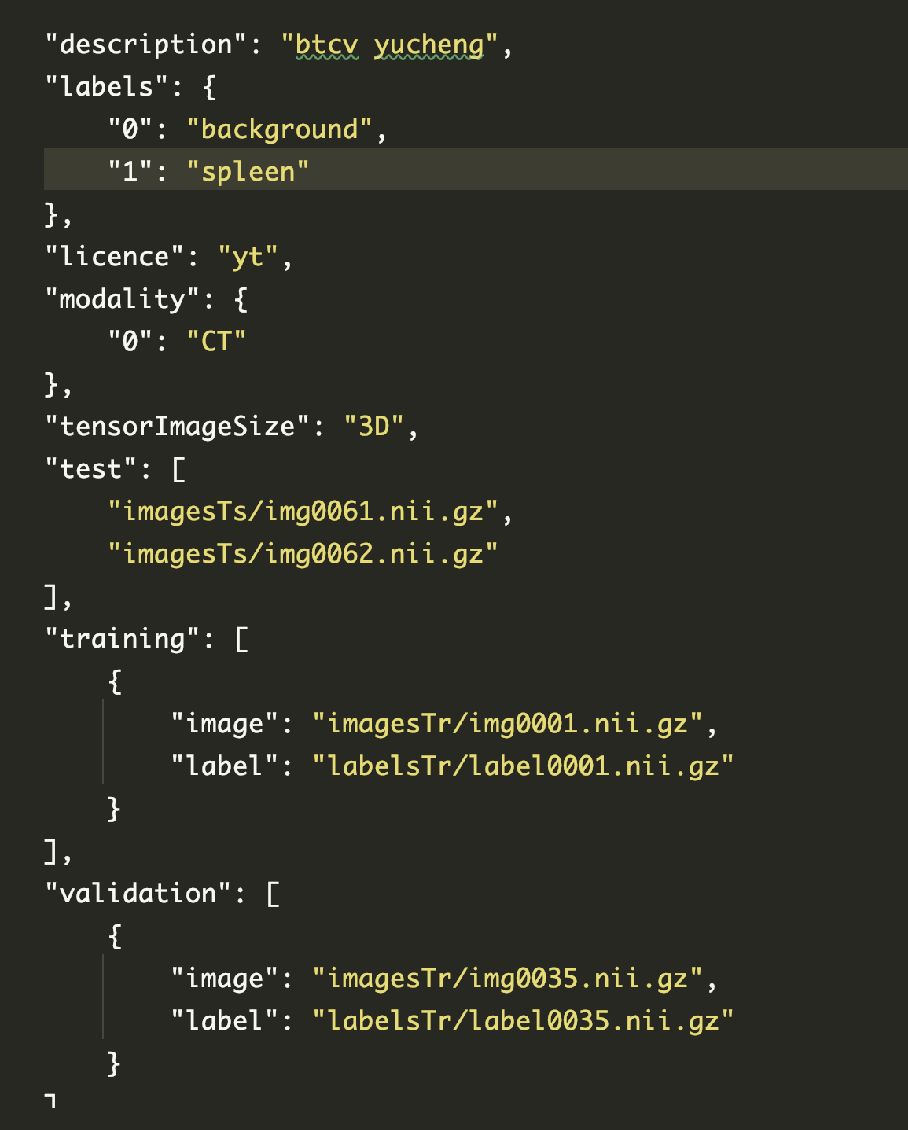
Copy the box and everything below to json file , Name it dataset_0.json
How to create json file : Baidu can , It's simple , ha-ha
structure Dataloader
data_dir = "data/"
split_JSON = "data/dataset_0.json"
# data_dir = "/home/ali/Desktop/data_local/Synapse_Orig/"
# split_JSON = "dataset_0.json"
datasets = data_dir + split_JSON
datalist = load_decathlon_datalist(datasets, True, "training")
val_files = load_decathlon_datalist(datasets, True, "validation")
train_ds = CacheDataset(
data=datalist,
transform=train_transforms,
cache_num=24,
cache_rate=1.0,
num_workers=8,
)
train_loader = DataLoader(
train_ds, batch_size=1, shuffle=True, num_workers=8, pin_memory=True
)
val_ds = CacheDataset(
data=val_files, transform=val_transforms, cache_num=6, cache_rate=1.0, num_workers=4
)
val_loader = DataLoader(
val_ds, batch_size=1, shuffle=False, num_workers=4, pin_memory=True
)
Here we need to pay attention to , Your data address should be written correctly .
Then run the next cell, Check whether the data is correct 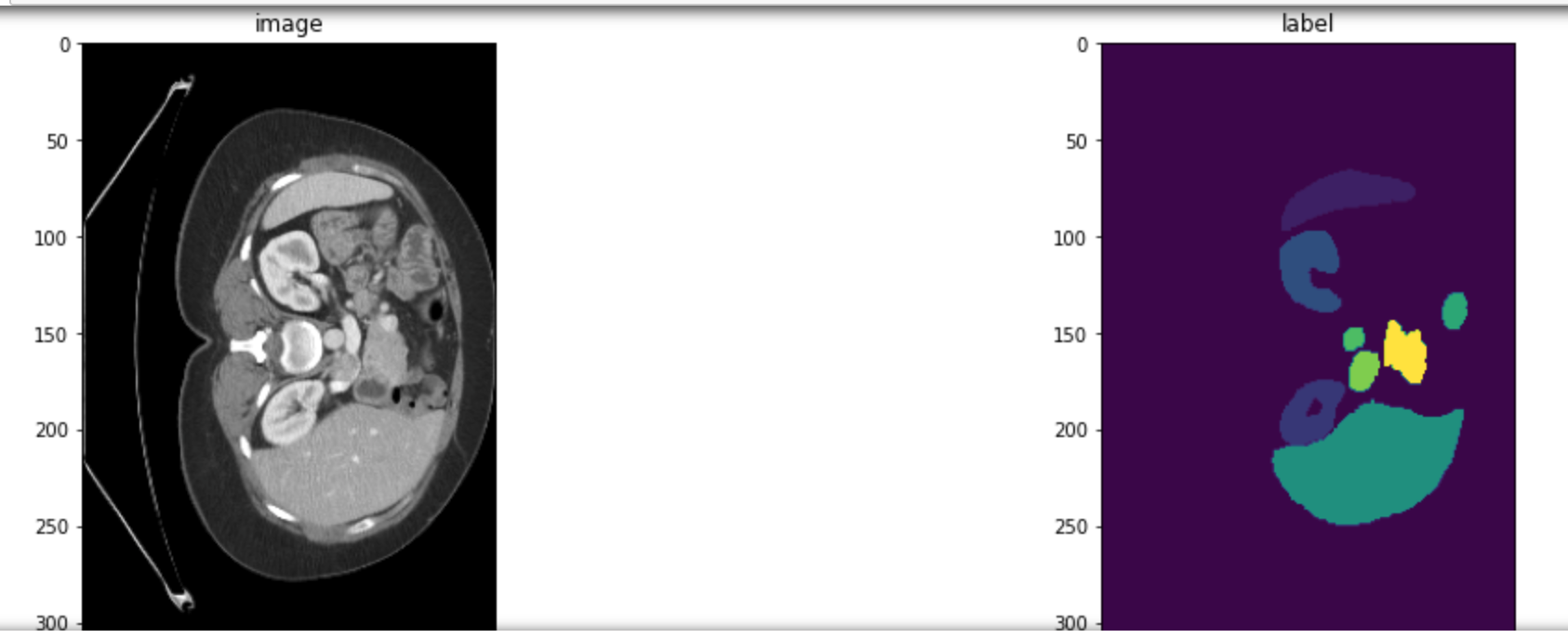
Build the model , Loss function , Optimizer
os.environ["CUDA_DEVICE_ORDER"] = "PCI_BUS_ID"
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model = UNETR(
in_channels=1,
out_channels=14,
img_size=(96, 96, 96),
feature_size=16,
hidden_size=768,
mlp_dim=3072,
num_heads=12,
pos_embed="perceptron",
norm_name="instance",
res_block=True,
dropout_rate=0.0,
).to(device)
loss_function = DiceCELoss(to_onehot_y=True, softmax=True)
torch.backends.cudnn.benchmark = True
optimizer = torch.optim.AdamW(model.parameters(), lr=1e-4, weight_decay=1e-5)
here , Is to put model Instead of UNETR, The rest are the same CNN There is no difference between .
Then there is a typical pytorch Training . The code is long , I won't paste it .
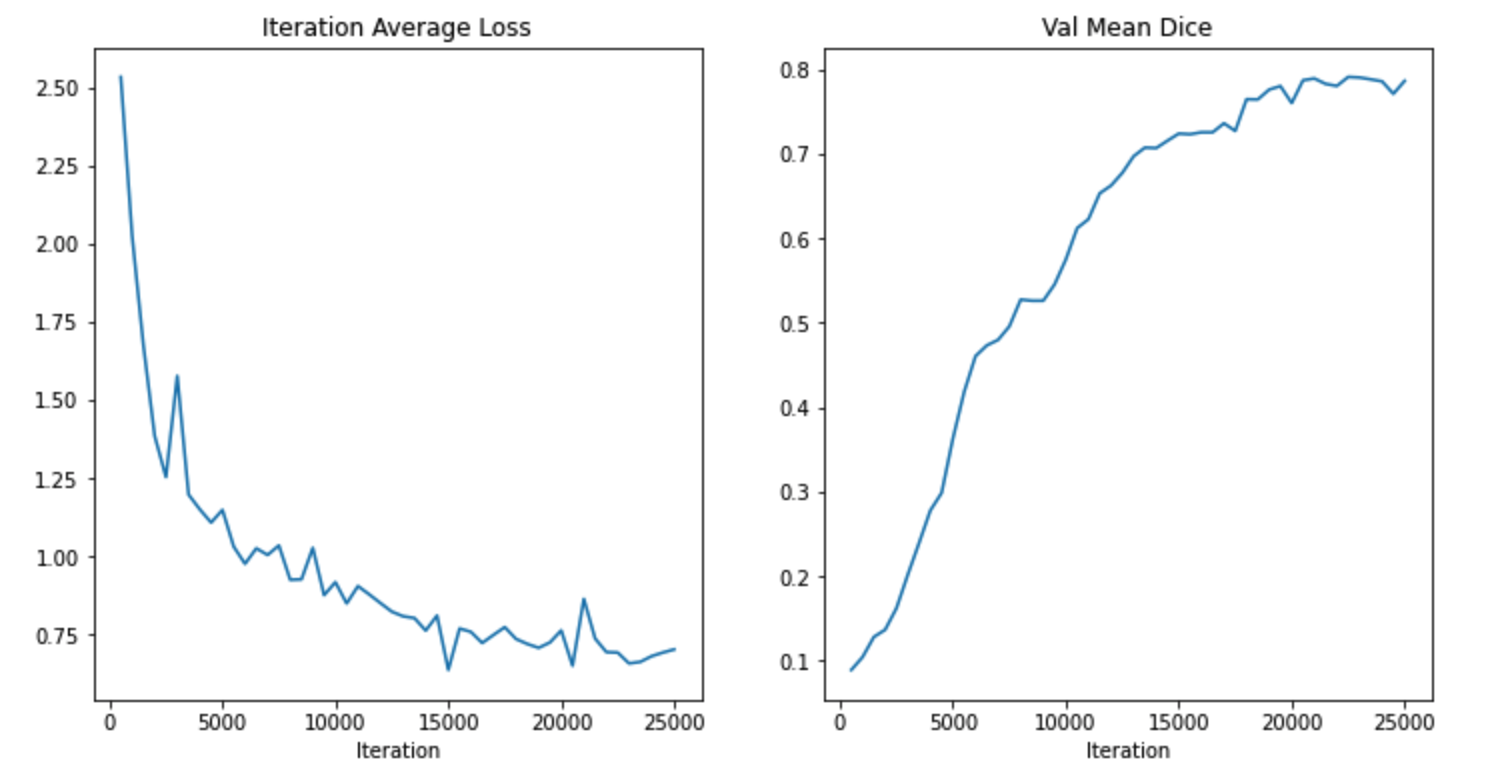
The result of my training is 0.7907, Not bad . It's also my first taste transformer The advantages of .
Let's try it quickly ~~
Articles are constantly updated , You can pay attention to the official account of WeChat 【 Medical image AI combat camp 】 Get the latest , The official account of the frontier technology in the field of medical image processing . Stick to the practice , Take you hand in hand to do the project , Play the game , Write a paper . All original articles provide theoretical explanation , Experimental code , experimental data . Only practice can grow faster , Pay attention to our , Learn together ~
I am a Tina, I'll see you on our next blog ~
Working during the day and writing at night , cough
If you think it's well written, finally , Please thumb up , Comment on , Collection . Or three times with one click 
边栏推荐
- Five simple and practical daily development functions of chrome are explained in detail. Unlock quickly to improve your efficiency!
- CC2530 zigbee IAR8.10.1环境搭建
- Multithreaded asynchronous orchestration
- P2788 math 1 - addition and subtraction
- 南航 PA3.1
- Installation and configuration of slurm resource management and job scheduling system
- 打算参加安全方面工作,信息安全工程师怎么样,软考考试需要怎么准备?
- [牛客网刷题 Day5] JZ77 按之字形顺序打印二叉树
- The width of table is 4PX larger than that of tbody
- [système recommandé 01] rechub
猜你喜欢
![[daiy5] jz77 print binary tree in zigzag order](/img/ba/b2dfbf121798757c7b9fba4811221b.png)
[daiy5] jz77 print binary tree in zigzag order

Experience sharing of software designers preparing for exams

Using U2 net deep network to realize -- certificate photo generation program
Multithreaded asynchronous orchestration
使用 load_decathlon_datalist (MONAI)快速加载JSON数据
![1323: [example 6.5] activity selection](/img/2e/ba74f1c56b8a180399e5d3172c7b6d.png)
1323: [example 6.5] activity selection
How to prepare for the advanced soft test (network planning designer)?
Leetcode-304: two dimensional area and retrieval - matrix immutable
XML configuration file parsing and modeling
Socket通信原理和实践
随机推荐
软考一般什么时候出成绩呢?在线蹬?
I'd rather say simple problems a hundred times than do complex problems once
P2788 数学1(math1)- 加减算式
深入理解Apache Hudi异步索引机制
@Transcation的配置,使用,原理注意事项:
MONAI版本更新到 0.9 啦,看看有什么新功能
打算参加安全方面工作,信息安全工程师怎么样,软考考试需要怎么准备?
About hzero resource error (groovy.lang.missingpropertyexception: no such property: weight for class)
深入理解Apache Hudi异步索引机制
Serial communication relay Modbus communication host computer debugging software tool project development case
Using U2 net deep network to realize -- certificate photo generation program
The width of table is 4PX larger than that of tbody
中级软件评测师考什么
SQL Server 知识汇集9 : 修改数据
Find the greatest common divisor and the least common multiple (C language)
Schnuka: machine vision positioning technology machine vision positioning principle
Sword finger offer 38 Arrangement of strings [no description written]
Study summary of postgraduate entrance examination in August
The mobile terminal automatically adjusts the page content and font size by setting rem
【PyTorch 07】 动手学深度学习——chapter_preliminaries/ndarray 习题动手版