当前位置:网站首页>[data clustering] section 3 of Chapter 4: DBSCAN performance analysis, advantages and disadvantages, and parameter selection methods
[data clustering] section 3 of Chapter 4: DBSCAN performance analysis, advantages and disadvantages, and parameter selection methods
2022-07-04 12:35:00 【Happy Jianghu】
List of articles
7、 ... and : Performance analysis
DBSCAN The algorithm retrieves all points in its neighborhood for each point in the data set , The time complexity is O ( n 2 ) O(n^{2}) O(n2). But in low latitude space , If the k k k- d d d Trees 、 R R R Trees and other structures , It can effectively retrieve all points within a given distance of a specific point , Its time complexity can be reduced to O ( n l o g n ) O(nlogn) O(nlogn)
8、 ... and : Advantages and disadvantages
(1) advantage
①: It can be done to Clustering dense data sets of arbitrary shape
②: We can find outliers while clustering , Insensitive to outliers in the dataset
③: There is no need to specify the number of clusters , And the results of many experiments are often the same
(2) shortcoming
①: If the density of the sample set is not uniform 、 When the distance between clusters is very different , The clustering quality is poor , Use DBSCAN Clustering is generally not suitable for
②: Tuning parameters are compared to K-Means More complicated , Different parameter combinations have a great impact on the clustering effect
③: Due to the traditional European distance Can't handle high latitude data well , Therefore, it is difficult to give a good solution to the definition of distance
④:DBSCAN The algorithm is suitable for dealing with the density of different clusters Relatively uniform The situation of , When When the density of different clusters changes greatly There will be some problems
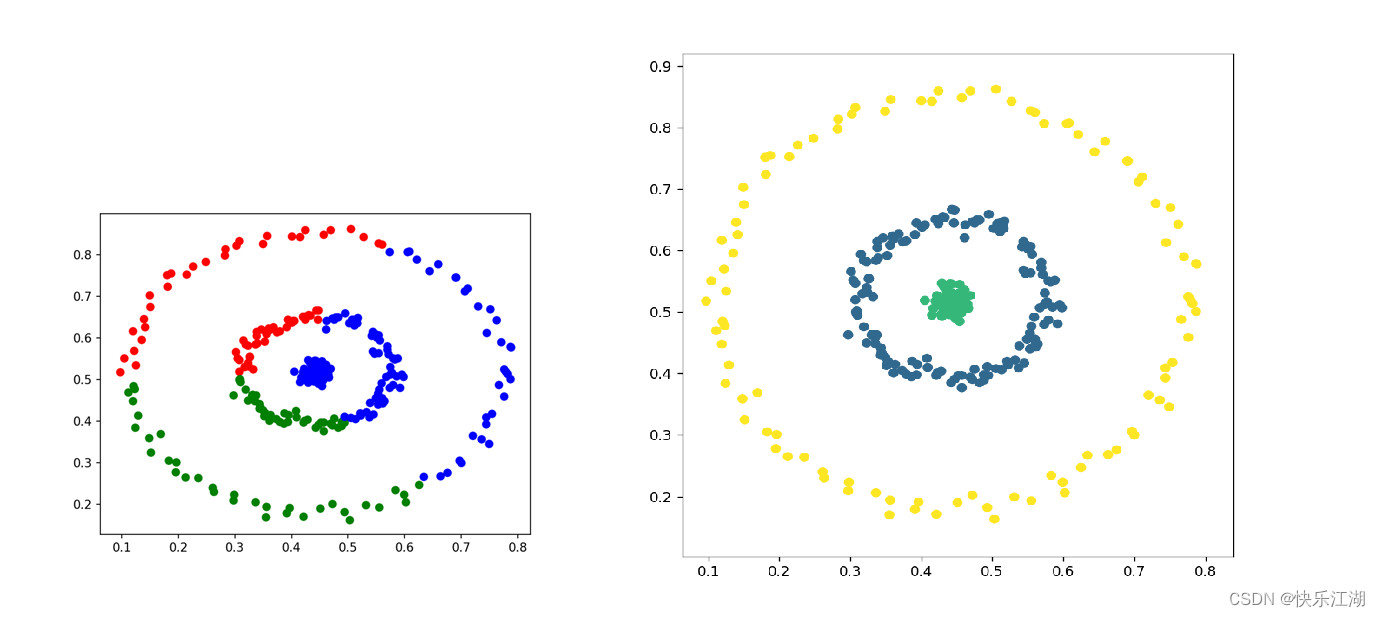
- From the clustering rendering shown in the previous section , You may find this algorithm It is easy to mark some data points as noise
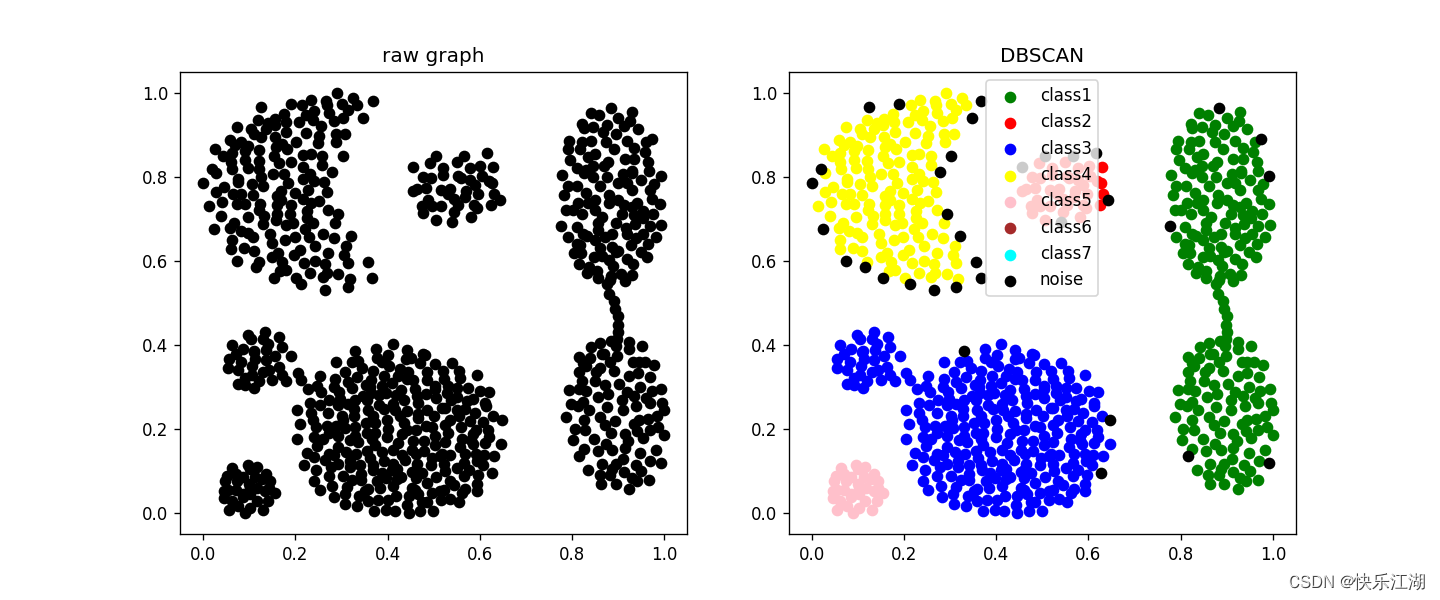
Nine : Parameter selection
(1) Basic principles of modifying parameters
When visualization is complete , We can fine tune the parameters according to the clustering results
min_pts: General choice dimension +1 Or dimension ×2 that will doeps: If you share Less classes , Then put eps Turn it down ; If you share There are more classes , Then put eps Turn it up
(2) according to K-dist Figure adjustment parameters
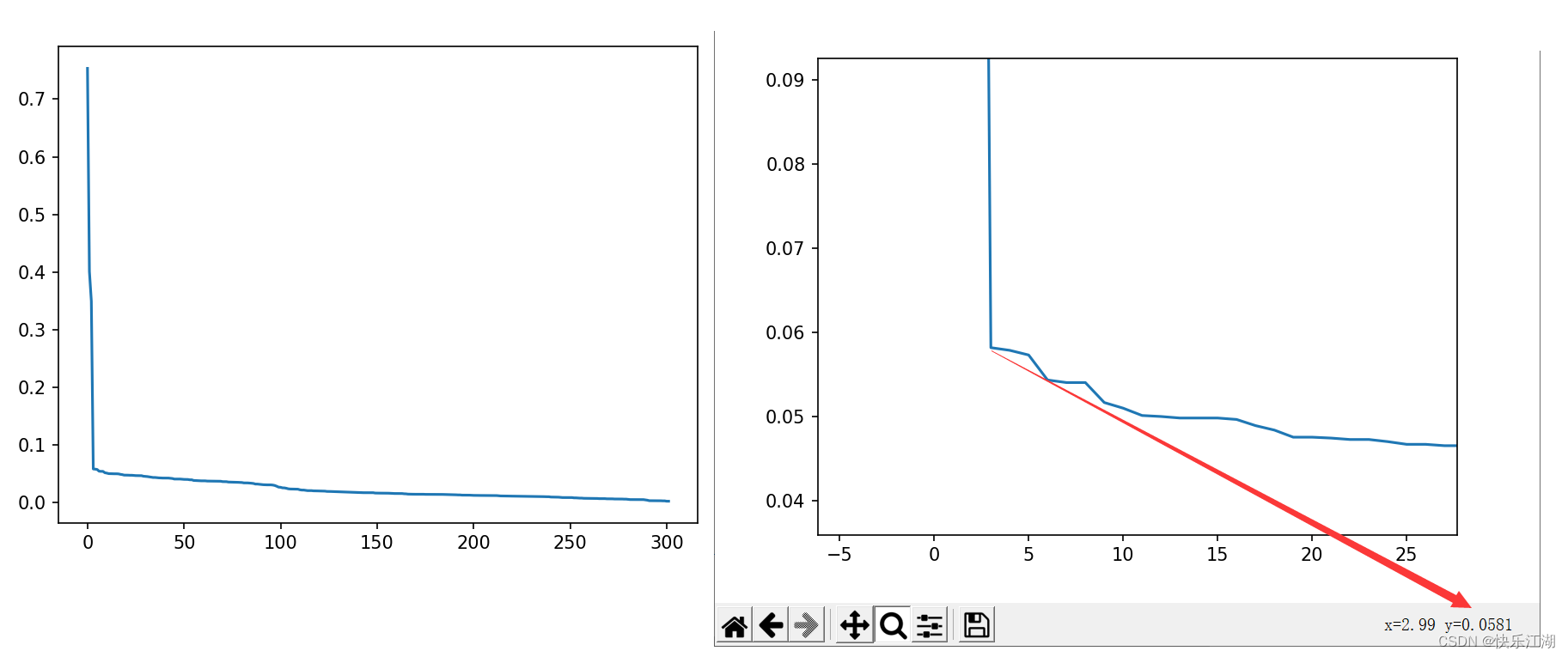
Parameter adjustment method : You can use observation The first k k k Nearest neighbor distance ( k k k distance ) Method to determine parameters . For a point in the cluster , If k k k No more than the number of samples in the cluster , be k k k The distance will be very small , then For noise , k k k The distance will be great . therefore : For any positive integer k k k, Calculate all data points k k k distance , Then sort it incrementally , Then draw k k k- Distance map , At the inflection point, it is appropriate eps, Because boundary points or noise points are often encountered at the inflection point
- k k k distance : For a given data set P P P= { p 1 , p 2 , . . . , p n } \{p_{1}, p_{2} , ... , p_{n}\} { p1,p2,...,pn}, Calculation point p ( i ) p(i) p(i) To dataset D D D Subset S S S The distance between all points in the , The distances are sorted from small to large , be d ( k ) d(k) d(k) It's called k k k distance
The specific steps are as follows
- determine K value , Usually take 2* dimension -1
- draw
K-distchart - Inflection point position ( The most volatile place ) by
eps( You can take multiple inflection points to try ) min_ptsCan take K+1
k = 3 # take 2*2-1
k_dist = select_minpts(train_data, k) # K Distance map
k_dist.sort() # Sort ( Default ascending order )
plt.plot(np.arange(k_dist.shape[0]), k_dist[::-1]) # Draw from large to small
plt.show()
def select_minpts(data_set, k):
k_dist = []
for i in range(data_set.shape[0]):
dist = (((data_set[i] - data_set)**2).sum(axis=1)**0.5)
dist.sort()
k_dist.append(dist[k])
return np.array(k_dist)
as follows eps It is suggested that 0.0581
Ten :DBSCAN Algorithm and K-Means Algorithm comparison
| DBSCAN | K-Means | |
|---|---|---|
| Algorithmic thought | be based on density | be based on Divide |
| Required parameters | Eps and MinPts | Number of clusters k k k |
| Data requirement | Make no assumptions about the data distribution | Suppose that all clusters come from a spherical Gaussian distribution |
| High dimensional data | Not suitable for | fit ( sparse ) |
| formalization | You can't | It can be formalized as an optimization problem that minimizes the sum of squares of errors from each point to the center of mass |
| stability | Stable | unstable |
| Time complexity | O ( n 2 ) O(n^{2}) O(n2) | O ( n ) O(n) O(n) |
边栏推荐
- What if the chat record is gone? How to restore wechat chat records on Apple Mobile
- World document to picture
- Possible to restore a backup of SQL Server 2014 on SQL Server 2012?
- [Yunju entrepreneurial foundation notes] Chapter II entrepreneur test 14
- The most robust financial products in 2022
- Error: Failed to download metadata for repo ‘AppStream‘: Cannot download repomd. XML solution
- C语言:求100-999是7的倍数的回文数
- [Yunju entrepreneurial foundation notes] Chapter II entrepreneur test 22
- The solution of permission denied
- [Yunju entrepreneurial foundation notes] Chapter II entrepreneur test 11
猜你喜欢
Awk getting started to proficient series - awk quick start
![[notes] in depth explanation of assets, resources and assetbundles](/img/e9/ae401b45743ea65986ae01b54e3593.jpg)
[notes] in depth explanation of assets, resources and assetbundles
Ml and NLP are still developing rapidly in 2021. Deepmind scientists recently summarized 15 bright research directions in the past year. Come and see which direction is suitable for your new pit
Practice of retro SOAP Protocol
Tableau makes data summary after linking the database, and summary exceptions occasionally occur.
Introduction to random and threadlocalrandom analysis
Source code analysis of the implementation mechanism of multisets in guava class library
![[Yunju entrepreneurial foundation notes] Chapter II entrepreneur test 16](/img/c3/f3746b161012acc3751b2bd0b8f663.jpg)
[Yunju entrepreneurial foundation notes] Chapter II entrepreneur test 16

The frost peel off the purple dragon scale, and the xiariba people will talk about database SQL optimization and the principle of indexing (primary / secondary / clustered / non clustered)
(August 10, 2021) web crawler learning - Chinese University ranking directed crawler
随机推荐
Star leap plan | new projects are continuously being recruited! MSR Asia MSR Redmond joint research program invites you to apply!
[Yunju entrepreneurial foundation notes] Chapter II entrepreneur test 11
Reptile learning 4 winter vacation learning series (1)
[Yunju entrepreneurial foundation notes] Chapter II entrepreneur test 19
13、 C window form technology and basic controls (3)
[Yunju entrepreneurial foundation notes] Chapter II entrepreneur test 18
Tableau makes data summary after linking the database, and summary exceptions occasionally occur.
Iterm tab switching order
Communication tutorial | overview of the first, second and third generation can bus
Complementary knowledge of auto encoder
How to realize the function of Sub Ledger of applet?
How do std:: function and function pointer assign values to each other
TCP fast retransmission sack mechanism
priority_ queue
Classification and application of AI chips
'using an alias column in the where clause in PostgreSQL' - using an alias column in the where clause in PostgreSQL
C language memory layout
Global and Chinese markets for environmental disinfection robots 2022-2028: Research Report on technology, participants, trends, market size and share
22 API design practices
[Yunju entrepreneurial foundation notes] Chapter II entrepreneur test 22