当前位置:网站首页>Label propagation
Label propagation
2022-07-02 07:39:00 【Xiao Chen who wants money】
Recently, I am studying the semi supervised algorithm of time series , See this algorithm , It was recorded .
Reprinted from : Tag propagation algorithm (Label Propagation) And Python Realization _zouxy09 The column -CSDN Blog _ Tag propagation algorithm
Semi-supervised learning (Semi-supervised learning) The occasion to play a role is : Some of your data have label, Some don't . And most of them don't , Only a few have label. Semi supervised learning algorithm will make full use of unlabeled Data to capture the potential distribution of our entire data . It is based on three assumptions :
1)Smoothness Smoothing hypothesis : Similar data have the same label.
2)Cluster Clustering hypothesis : Data under the same cluster have the same label.
3)Manifold Manifold hypothesis : Data in the same manifold structure have the same label.
Tag propagation algorithm (label propagation) The core idea of is very simple : Similar data should have the same label.LP The algorithm consists of two steps :1) Construct similarity matrix (affinity matrix);2) Spread it bravely .
label propagation Is a graph based algorithm . Graph is based on vertices and edges , Each vertex is a sample , All vertices include labeled samples and unlabeled samples ; Edges represent vertices i To the top j Probability , In other words, the vertex i To the top j The similarity .

here ,α It's a superscript .
Another very common method of graph construction is knn chart , That is, only for each node k Nearest neighbor weight , The other is 0, That is, there is no edge , So it's a sparse similarity matrix .
The label propagation algorithm is very simple : Propagate through the edges between nodes label. The more weight an edge has , Indicates that the more similar two nodes are , that label The easier it is to spread the past . Let's define a NxN Probability transition matrix P:
Pij Represents slave node i Transfer to node j Probability . Suppose there is C Class and L individual labeled sample , Let's define a LxC Of label matrix YL, The first i Line representation i Labels of samples indicate vectors , That is, if the second i The categories of samples are j, Then the No j Elements are 1, For the other 0. Again , We also give U individual unlabeled One sample UxC Of label matrix YU. Merge them , We get a NxC Of soft label matrix F=[YL;YU].soft label It means , We keep the sample i The probability of belonging to each category , Not mutually exclusive , This sample is based on probability 1 Only belong to one class . Yes, of course , Finally determine this sample i Of the category , Is to take max That is, the class with the greatest probability is its class . that F There's a YU, It didn't know at first , What is the initial value ? It doesn't matter , Just set any value .
Come out , ordinary LP Algorithm is as follows :
1) Execute propagation :F=PF
2) Reset F in labeled The label of the sample :FL=YL
3) Repeat step 1) and 2) until F convergence .
step 1) That's to put the matrix P And matrices F Multiply , This step , Each node will have its own label With P The determined probability is propagated to other nodes . If the two nodes are more similar ( The closer the distance in European space ), So the other side's label The easier it is to be label give , It's easier to form gangs . step 2) It's critical , because labeled Data label It's predetermined , It cannot be taken away , So after each transmission , It has to return to its original label. With labeled The data will continue to own label Spread it out , The last class boundary will cross the high-density region , And stay in low-density intervals . Equivalent to each different category labeled The sample divides the sphere of influence .
2.3、 Transformed LP Algorithm
We know , We calculate one for each iteration soft label matrix F=[YL;YU], however YL It is known. , It's useless to calculate it , Steps in 2) When , We have to get it back . All we care about is YU, Can we just calculate YU Well ?Yes. We will matrix P Make the following division :

Now , Our algorithm is just an operation :

Iterate the above steps until convergence ok 了 , Isn't it very cool. You can see FU Not only depends on labeled Label of data and its transfer probability , It depends unlabeled Current data label And transition probability . therefore LP The algorithm can be used additionally unlabeled Distribution characteristics of data .
The convergence of this algorithm is also very easy to prove , See references for details [1]. actually , It can converge to a convex solution :

So we can also solve it directly in this way , To get the final YU. But in the actual application process , Because matrix inversion requires O(n3) Complexity , So if unlabeled There's a lot of data , that I – PUU The inversion of the matrix will be very time-consuming , Therefore, at this time, we usually choose iterative algorithm to realize .
#***************************************************************************
#*
#* Description: label propagation
#* Author: Zou Xiaoyi ([email protected])
#* Date: 2015-10-15
#* HomePage: http://blog.csdn.net/zouxy09
#*
#**************************************************************************
import time
import numpy as np
# return k neighbors index
def navie_knn(dataSet, query, k):
numSamples = dataSet.shape[0]
## step 1: calculate Euclidean distance
diff = np.tile(query, (numSamples, 1)) - dataSet
squaredDiff = diff ** 2
squaredDist = np.sum(squaredDiff, axis = 1) # sum is performed by row
## step 2: sort the distance
sortedDistIndices = np.argsort(squaredDist)
if k > len(sortedDistIndices):
k = len(sortedDistIndices)
return sortedDistIndices[0:k]
# build a big graph (normalized weight matrix)
def buildGraph(MatX, kernel_type, rbf_sigma = None, knn_num_neighbors = None):
num_samples = MatX.shape[0]
affinity_matrix = np.zeros((num_samples, num_samples), np.float32)
if kernel_type == 'rbf':
if rbf_sigma == None:
raise ValueError('You should input a sigma of rbf kernel!')
for i in xrange(num_samples):
row_sum = 0.0
for j in xrange(num_samples):
diff = MatX[i, :] - MatX[j, :]
affinity_matrix[i][j] = np.exp(sum(diff**2) / (-2.0 * rbf_sigma**2))
row_sum += affinity_matrix[i][j]
affinity_matrix[i][:] /= row_sum
elif kernel_type == 'knn':
if knn_num_neighbors == None:
raise ValueError('You should input a k of knn kernel!')
for i in xrange(num_samples):
k_neighbors = navie_knn(MatX, MatX[i, :], knn_num_neighbors)
affinity_matrix[i][k_neighbors] = 1.0 / knn_num_neighbors
else:
raise NameError('Not support kernel type! You can use knn or rbf!')
return affinity_matrix
# label propagation
def labelPropagation(Mat_Label, Mat_Unlabel, labels, kernel_type = 'rbf', rbf_sigma = 1.5, \
knn_num_neighbors = 10, max_iter = 500, tol = 1e-3):
# initialize
num_label_samples = Mat_Label.shape[0]
num_unlabel_samples = Mat_Unlabel.shape[0]
num_samples = num_label_samples + num_unlabel_samples
labels_list = np.unique(labels)
num_classes = len(labels_list)
MatX = np.vstack((Mat_Label, Mat_Unlabel))
clamp_data_label = np.zeros((num_label_samples, num_classes), np.float32)
for i in xrange(num_label_samples):
clamp_data_label[i][labels[i]] = 1.0
label_function = np.zeros((num_samples, num_classes), np.float32)
label_function[0 : num_label_samples] = clamp_data_label
label_function[num_label_samples : num_samples] = -1
# graph construction
affinity_matrix = buildGraph(MatX, kernel_type, rbf_sigma, knn_num_neighbors)
# start to propagation
iter = 0; pre_label_function = np.zeros((num_samples, num_classes), np.float32)
changed = np.abs(pre_label_function - label_function).sum()
while iter < max_iter and changed > tol:
if iter % 1 == 0:
print "---> Iteration %d/%d, changed: %f" % (iter, max_iter, changed)
pre_label_function = label_function
iter += 1
# propagation
label_function = np.dot(affinity_matrix, label_function)
# clamp
label_function[0 : num_label_samples] = clamp_data_label
# check converge
changed = np.abs(pre_label_function - label_function).sum()
# get terminate label of unlabeled data
unlabel_data_labels = np.zeros(num_unlabel_samples)
for i in xrange(num_unlabel_samples):
unlabel_data_labels[i] = np.argmax(label_function[i+num_label_samples])
return unlabel_data_labels边栏推荐
- 【BERT,GPT+KG调研】Pretrain model融合knowledge的论文集锦
- Drawing mechanism of view (3)
- label propagation 标签传播
- Generate random 6-bit invitation code in PHP
- 基于onnxruntime的YOLOv5单张图片检测实现
- A slide with two tables will help you quickly understand the target detection
- MySQL has no collation factor of order by
- Record of problems in the construction process of IOD and detectron2
- 【Torch】最简洁logging使用指南
- mmdetection训练自己的数据集--CVAT标注文件导出coco格式及相关操作
猜你喜欢
SSM laboratory equipment management
MySQL has no collation factor of order by

Traditional target detection notes 1__ Viola Jones
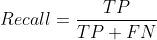
Common machine learning related evaluation indicators

iOD及Detectron2搭建过程问题记录

MoCO ——Momentum Contrast for Unsupervised Visual Representation Learning
Implementation of yolov5 single image detection based on onnxruntime
论文写作tip2
![[medical] participants to medical ontologies: Content Selection for Clinical Abstract Summarization](/img/24/09ae6baee12edaea806962fc5b9a1e.png)
[medical] participants to medical ontologies: Content Selection for Clinical Abstract Summarization

ERNIE1.0 与 ERNIE2.0 论文解读
随机推荐
Calculate the difference in days, months, and years between two dates in PHP
自然辩证辨析题整理
机器学习理论学习:感知机
生成模型与判别模型的区别与理解
yolov3训练自己的数据集(MMDetection)
MMDetection模型微调
ModuleNotFoundError: No module named ‘pytest‘
Record of problems in the construction process of IOD and detectron2
Interpretation of ernie1.0 and ernie2.0 papers
How to efficiently develop a wechat applet
PointNet原理证明与理解
Faster-ILOD、maskrcnn_ Benchmark training coco data set and problem summary
Typeerror in allenlp: object of type tensor is not JSON serializable error
Alpha Beta Pruning in Adversarial Search
使用Matlab实现:弦截法、二分法、CG法,求零点、解方程
Calculate the total in the tree structure data in PHP
Machine learning theory learning: perceptron
[paper introduction] r-drop: regulated dropout for neural networks
Get the uppercase initials of Chinese Pinyin in PHP
ABM thesis translation