当前位置:网站首页>Summary of anomaly detection methods
Summary of anomaly detection methods
2022-07-06 06:12:00 【Zhan Miao】
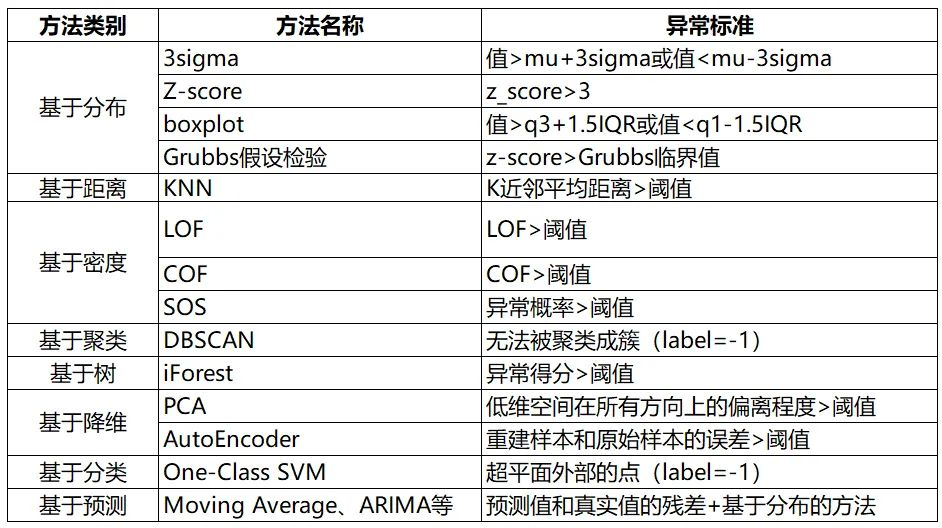
1. Distribution based approach
1.1. 3sigma
Based on the normal distribution ,3sigma The code says that it exceeds 3sigma The data of is outliers .

def three_sigma(s):
mu, std = np.mean(s), np.std(s)
lower, upper = mu-3*std, mu+3*std
return lower, upper1.2. Z-score
Z-score Is the standard score , Measure the distance between the data point and the average value , if A The difference from the average is 2 A standard deviation ,Z-score by 2. When put Z-score=3 As a threshold to eliminate outliers , It is equivalent to 3sigma.
def z_score(s):
z_score = (s - np.mean(s)) / np.std(s)
return z_score1.3. boxplot
The boxplot is based on the interquartile distance (IQR) Looking for outliers .

def boxplot(s):
q1, q3 = s.quantile(.25), s.quantile(.75)
iqr = q3 - q1
lower, upper = q1 - 1.5*iqr, q3 + 1.5*iqr
return lower, upper1.4. Grubbs Hypothesis testing
Grubbs’Test It is a method of hypothesis testing , It is often used to test univariate data sets with normal distribution (univariate data set)Y Single outliers in . If there are abnormal values , It must be the maximum or minimum value in the data set . The original and alternative assumptions are as follows :
H0: There are no outliers in the dataset
H1: There is an outlier in the dataset
Use Grubbs Testing requires that the population be normally distributed . Algorithm flow :
1. The samples are sorted from small to large
2. For the sample mean and dev
3. Calculation min/max And mean The gap between , The bigger one is suspicious
4. For suspicious values z-score (standard score), If it is greater than Grubbs critical value , So that is outlier
Grubbs The critical value can be obtained by looking up the table , It's determined by two values : detection level α( The more strict, the smaller ), Number of samples n, exclude outlier, Do... For the remaining sequence loop 1-4 step [1]. For detailed calculation examples, please refer to .
from outliers import smirnov_grubbs as grubbs
print(grubbs.test([8, 9, 10, 1, 9], alpha=0.05))
print(grubbs.min_test_outliers([8, 9, 10, 1, 9], alpha=0.05))
print(grubbs.max_test_outliers([8, 9, 10, 1, 9], alpha=0.05))
print(grubbs.max_test_indices([8, 9, 10, 50, 9], alpha=0.05))limited :
- Only single dimension data can be detected
- Unable to accurately output the normal interval
- Its judging mechanism is “ Eliminate one by one ”, So each outlier should be calculated separately for the whole step , Too much data .
- It is necessary to assume that the data obey normal distribution or near normal distribution
2. Distance based approach
2.1. KNN
Calculate each sample point and its nearest... In turn K The average distance between samples , Then compare the calculated distance with the threshold , If it is greater than the threshold , It is considered to be an outlier . The advantage is that there is no need to assume the distribution of data , The disadvantage is that only global outliers can be found , Local outliers cannot be found .
from pyod.models.knn import KNN
# Initialize detector clf
clf = KNN( method='mean', n_neighbors=3, )
clf.fit(X_train)
# Return the classification label on the training data (0: Normal value , 1: outliers )
y_train_pred = clf.labels_
# Return outliers on training data ( The higher the score, the more abnormal )
y_train_scores = clf.decision_scores_3. Density based approach
3.1. Local Outlier Factor (LOF)
LOF It's a classic density based algorithm (Breuning et. al. 2000), By assigning an outlier factor that depends on the neighborhood density to each data point LOF, Then judge whether the data point is an outlier . Its advantage is that it can quantify the abnormal degree of each data point (outlierness).

The data points p The local relative density of ( Local anomaly factor ) Is a point p The average local reachable density of points in the neighborhood is the same as that of data points p The ratio of the locally attainable density of , namely :
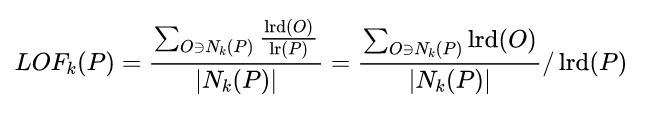
The data points P Of Local accessible density =P The reciprocal of the average reachable distance of the nearest neighbor . The greater the distance , The smaller the density .
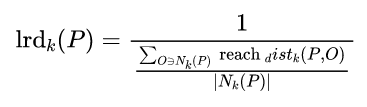
spot P point-to-point O Of The first k Reach distance =max( spot O Of k Neighbor distance , spot P point-to-point O Distance of ).
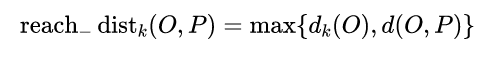
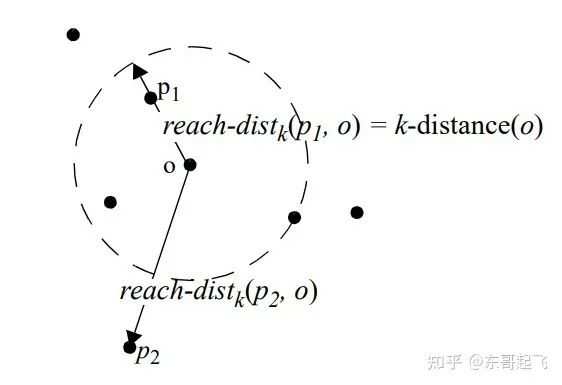
spot O Of k Neighbor distance = The first k The nearest point and point O Distance between .
As a whole ,LOF The algorithm flow is as follows :
For each data point , Calculate its distance from all the other points , And sort it from near to far ;
For each data point , Find it. K-Nearest-Neighbor, Calculation LOF score .
from sklearn.neighbors import LocalOutlierFactor as LOF
X = [[-1.1], [0.2], [100.1], [0.3]]
clf = LOF(n_neighbors=2)
res = clf.fit_predict(X)
print(res)
print(clf.negative_outlier_factor_)3.2. Connectivity-Based Outlier Factor (COF)
COF yes LOF Variants , Compared with LOF,COF It can handle outliers in low density ,COF The local density of is calculated based on the average chain distance . At the beginning, we will also calculate the value of each point k-nearest neighbor. And then we'll calculate for each point Set based nearest Path, Here's the picture :

If we today our k=5, therefore F Of neighbor by B、C、D、E、G. And for F The nearest point to him is E, therefore SBN Path The first element of is F、 The second is E. leave E The nearest point is D So the third element is D, Next, leave D The nearest point is C and G, So the fourth and fifth elements are C and G, Finally leave C The nearest point is B, The sixth element is B. So the whole process goes down ,F Of SBN Path by {F, E, D, C, G, C, B}. And for SBN Path The corresponding distance e={e1, e2, e3,…,ek}, According to the example above e={3,2,1,1,1}.
So we can say that if we want to calculate p Dot SBN Path, We just need to calculate directly p Point and its neighbor Of all the points graph Of minimum spanning tree, Then we'll take p Point as the starting point shortest path Algorithm , You can get our SBN Path.
And then we have SBN Path We will then calculate ,p Chain distance of points :
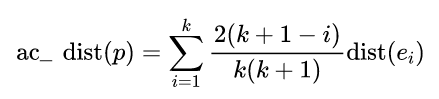
With ac_distance after , We can calculate COF:
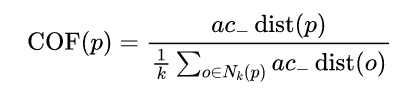
from pyod.models.cof import COF
cof = COF(contamination = 0.06, ## The proportion of outliers
n_neighbors = 20, ## Number of nearest neighbors
)
cof_label = cof.fit_predict(iris.values) # Iris data
print(" The number of outliers detected is :",np.sum(cof_label == 1))3.3. Stochastic Outlier Selection (SOS)
The characteristic matrix (feature martrix) Or dissimilarity matrix (dissimilarity matrix) Input to SOS Algorithm , Will return an abnormal probability value vector ( Each point corresponds to a ).SOS The idea is : When a point is related to all other points (affinity) When they were very young , It is an outlier .

SOS The process of :
- Calculate the dissimilarity matrix D;
- Calculate the incidence matrix A;
- Calculate the incidence probability matrix B;
- Calculate the anomaly probability vector .
Dissimilarity matrix D Is the measured distance between two samples , Such as European distance or Hamming distance . The incidence matrix reflects Measure distance variance , Pictured 7, spot  Has the highest density , Minimum variance ;
Has the highest density , Minimum variance ;  Has the lowest density , The largest variance . And the incidence probability matrix B
Has the lowest density , The largest variance . And the incidence probability matrix B
(binding probability matrix) The incidence matrix (affinity matrix) Normalized by rows , Pictured 8 Shown .


Got it binding probability matrix, The abnormal probability value of each point is calculated by the following formula , When a point is related to all other points (affinity) When they were very young , It is an outlier .
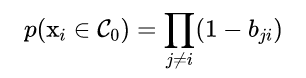
import pandas as pd
from sksos import SOS
iris = pd.read_csv("http://bit.ly/iris-csv")
X = iris.drop("Name", axis=1).values
detector = SOS()
iris["score"] = detector.predict(X)
iris.sort_values("score", ascending=False).head(10)4. Clustering based methods
4.1. DBSCAN
DBSCAN Algorithm (Density-Based Spatial Clustering of Applications with Noise) The inputs and outputs of are as follows , For outliers that cannot form clusters , This is the outlier ( Noise point ).
Input : Data sets , Neighborhood radius Eps, Threshold for the number of data objects in the neighborhood MinPts;
Output : Density connected cluster .

The process is as follows :
- Select any data object point from the data set p;
- If for parameters Eps and MinPts, Selected data object point p As the core point , Then find out all from p Density up to data object points , Form a cluster ;
- If the selected data object point p It's the edge point , Select another data object point ;
- Repeat above 2、3 Step , Until all points are processed .
from sklearn.cluster import DBSCAN
import numpy as np
X = np.array([[1, 2], [2, 2], [2, 3],
[8, 7], [8, 8], [25, 80]])
clustering = DBSCAN(eps=3, min_samples=2).fit(X)
clustering.labels_
array([ 0, 0, 0, 1, 1, -1])
# 0,,0,,0: Indicates that the first three samples are divided into a group
# 1, 1: The middle two are divided into a group
# -1: The last one is the outlier , Not belonging to any group 5. Tree based approach
5.1. Isolation Forest (iForest)
Isolated forest “ isolated ” (isolation) refer to “ Isolate outliers from all samples ”, The original text in the paper is “separating an instance from the rest of the instances”.
We use a random hyperplane to cut a data space , You can generate two subspaces at a time . Next , Let's continue to randomly select hyperplanes , To cut the two subspaces obtained in the first step , It's a cycle , Until each subspace contains only one data point . We can find out , Those clusters with high density have to be cut many times before they stop cutting , That is, each point exists separately in a subspace , But those sparse points , Most of them stopped in a subspace very early . therefore , The algorithm idea of the whole isolated forest : Abnormal samples are more likely to fall into leaf nodes or , The abnormal sample is on the decision tree , Closer to the root node .
Random selection m Features , By randomly selecting a value between the maximum value and the minimum value of the selected feature to segment the data points . The partition of observations repeats recursively , Until all observations are isolated .

get t After an isolated tree , The training of a single tree is over . Then you can use the generated isolated tree to evaluate the test data , Calculate the abnormal score s. For each sample x, It is necessary to comprehensively calculate the results of each tree , Calculate the abnormal score by the following formula :

from sklearn.datasets import load_iris
from sklearn.ensemble import IsolationForest
data = load_iris(as_frame=True)
X,y = data.data,data.target
df = data.frame
# model training
iforest = IsolationForest(n_estimators=100, max_samples='auto',
contamination=0.05, max_features=4,
bootstrap=False, n_jobs=-1, random_state=1)
# fit_predict function Training and prediction work together Whether the abnormal model can be judged ,-1 Is abnormal ,1 It's normal
df['label'] = iforest.fit_predict(X)
# forecast decision_function We can draw Abnormal score
df['scores'] = iforest.decision_function(X)6. Method based on dimension reduction
6.1. Principal Component Analysis (PCA)
PCA The practice of anomaly detection , There are generally two ideas :
(1) Mapping data to low dimensional feature space , Then we can check the deviation between each data point and other data in different dimensions of feature space ;
(2) Mapping data to low dimensional feature space , Then the low dimensional feature space is mapped back to the original space , Try to reconstruct the original data with low dimensional features , Look at the size of the reconstruction error .
PCA Doing eigenvalue decomposition , You'll get :
- Eigenvector : It reflects the different directions of variance variation of original data ;
- The eigenvalue : The variance of the data in the corresponding direction .

The score is greater than the threshold C Then it is judged as abnormal .
The second way ,PCA The main features of the data are extracted , If a data sample is not easy to reconstruct , Indicates that the characteristics of this data sample are inconsistent with those of the overall data sample , So it's obviously an abnormal sample :

When reconstructing data samples based on low dimensional features , The information in the direction of the eigenvector corresponding to the smaller eigenvalue is discarded . In other words , In fact, the reconstruction error mainly comes from the information in the direction of the eigenvector corresponding to the smaller eigenvalues . Based on this intuitive understanding ,PCA The two different approaches to anomaly detection pay special attention to the eigenvectors corresponding to smaller eigenvalues . therefore , We said PCA The two approaches to anomaly detection are essentially similar , Of course, the first method can also focus on the eigenvectors corresponding to larger eigenvalues .
from sklearn.decomposition import PCA
pca = PCA()
pca.fit(centered_training_data)
transformed_data = pca.transform(training_data)
y = transformed_data
# Calculate the exception score
lambdas = pca.singular_values_
M = ((y*y)/lambdas)
# front k Eigenvectors and post r eigenvectors
q = 5
print "Explained variance by first q terms: ", sum(pca.explained_variance_ratio_[:q])
q_values = list(pca.singular_values_ < .2)
r = q_values.index(True)
# Calculate the sum of distances for each sample point
major_components = M[:,range(q)]
minor_components = M[:,range(r, len(features))]
major_components = np.sum(major_components, axis=1)
minor_components = np.sum(minor_components, axis=1)
# Artificially set c1、c2 threshold
components = pd.DataFrame({'major_components': major_components,
'minor_components': minor_components})
c1 = components.quantile(0.99)['major_components']
c2 = components.quantile(0.99)['minor_components']
# Make classifiers
def classifier(major_components, minor_components):
major = major_components > c1
minor = minor_components > c2
return np.logical_or(major,minor)
results = classifier(major_components=major_components, minor_components=minor_components)6.2. AutoEncoder
PCA It is linear dimensionality reduction ,AutoEncoder Is a nonlinear dimensionality reduction . Trained according to normal data AutoEncoder, It can reconstruct and restore normal samples , However, it is impossible to restore data points different from the normal distribution , Resulting in large reduction error . So if a new sample is encoded , After decoding , Its error exceeds the error range after normal data encoding and decoding , It is regarded as abnormal data . It should be noted that ,AutoEncoder The data used for training is normal data ( That is, there is no abnormal value ), Only in this way can we get the reasonable and normal error distribution range after reconstruction . therefore AutoEncoder When doing anomaly detection here , It is a method of supervised learning .

import tensorflow as tf
from keras.models import Sequential
from keras.layers import Dense
# Standardized data
scaler = preprocessing.MinMaxScaler()
X_train = pd.DataFrame(scaler.fit_transform(dataset_train),
columns=dataset_train.columns,
index=dataset_train.index)
# Random shuffle training data
X_train.sample(frac=1)
X_test = pd.DataFrame(scaler.transform(dataset_test),
columns=dataset_test.columns,
index=dataset_test.index)
tf.random.set_seed(10)
act_func = 'relu'
# Input layer:
model=Sequential()
# First hidden layer, connected to input vector X.
model.add(Dense(10,activation=act_func,
kernel_initializer='glorot_uniform',
kernel_regularizer=regularizers.l2(0.0),
input_shape=(X_train.shape[1],)
)
)
model.add(Dense(2,activation=act_func,
kernel_initializer='glorot_uniform'))
model.add(Dense(10,activation=act_func,
kernel_initializer='glorot_uniform'))
model.add(Dense(X_train.shape[1],
kernel_initializer='glorot_uniform'))
model.compile(loss='mse',optimizer='adam')
print(model.summary())
# Train model for 100 epochs, batch size of 10:
NUM_EPOCHS=100
BATCH_SIZE=10
history=model.fit(np.array(X_train),np.array(X_train),
batch_size=BATCH_SIZE,
epochs=NUM_EPOCHS,
validation_split=0.05,
verbose = 1)
plt.plot(history.history['loss'],
'b',
label='Training loss')
plt.plot(history.history['val_loss'],
'r',
label='Validation loss')
plt.legend(loc='upper right')
plt.xlabel('Epochs')
plt.ylabel('Loss, [mse]')
plt.ylim([0,.1])
plt.show()
# See how the error distribution of the training set restore , In order to establish the normal error distribution range
X_pred = model.predict(np.array(X_train))
X_pred = pd.DataFrame(X_pred,
columns=X_train.columns)
X_pred.index = X_train.index
scored = pd.DataFrame(index=X_train.index)
scored['Loss_mae'] = np.mean(np.abs(X_pred-X_train), axis = 1)
plt.figure()
sns.distplot(scored['Loss_mae'],
bins = 10,
kde= True,
color = 'blue')
plt.xlim([0.0,.5])
# Error threshold comparison , Find outliers
X_pred = model.predict(np.array(X_test))
X_pred = pd.DataFrame(X_pred,
columns=X_test.columns)
X_pred.index = X_test.index
threshod = 0.3
scored = pd.DataFrame(index=X_test.index)
scored['Loss_mae'] = np.mean(np.abs(X_pred-X_test), axis = 1)
scored['Threshold'] = threshod
scored['Anomaly'] = scored['Loss_mae'] > scored['Threshold']
scored.head()7. A classification based approach
7.1. One-Class SVM
One-Class SVM, The idea of this algorithm is very simple , Is to find a hyperplane to circle the positive examples in the sample , Prediction is to use this hyperplane to make decisions , A sample in a circle is considered a positive sample , The sample outside the circle is a negative sample , Used in anomaly detection , Negative samples can be regarded as abnormal samples . It belongs to unsupervised learning , So you don't need labels .

One-Class SVM Another way of deriving is SVDD(Support Vector Domain Description, Support vector domain description ), about SVDD Come on , We expect all samples that are not abnormal to be positive , And it uses a hypersphere , Instead of using a hyperplane to partition , The algorithm obtains the spherical boundary around the data in the feature space , I want to minimize the volume of this hypersphere , To minimize the impact of outlier data .

C Is to adjust the influence of relaxation variables , To put it mildly , How much slack space for data points that need to be relaxed , If C The relatively small , Will give the outliers greater flexibility , So that they can not be included in the hypersphere .
from sklearn import svm
# fit the model
clf = svm.OneClassSVM(nu=0.1, kernel='rbf', gamma=0.1)
clf.fit(X)
y_pred = clf.predict(X)
n_error_outlier = y_pred[y_pred == -1].size8. Prediction based approach
For single time series data , The predicted time series curve is compared with the real data , Find the residuals for each point , And the residual sequence is modeled , utilize KSigma Or quantile and other methods can be used for anomaly detection . The specific process is as follows :

9. summary
The anomaly detection methods are summarized as follows :

Reference material
[1] An overview of anomaly detection algorithms for time series prediction competitions - Fish in the rain , You know :https://zhuanlan.zhihu.com/p/336944097
[2] Weeding outliers grid calculator _ Statistics needed by data analysts : Anomaly detection - weixin_39974030,CSDN:https://blog.csdn.net/weixin_39974030/article/details/112569610
[3] Anomaly detection algorithm (KNN)-K Nearest Neighbors - Brother Wu talks about risk control , You know :https://zhuanlan.zhihu.com/p/501691799
[4] Read and understand anomaly detection LOF Algorithm (Python Code )- Dongge takes off , You know :https://zhuanlan.zhihu.com/p/448276009
[5] Nowak-Brzezińska, A., & Horyń, C. (2020). Outliers in rules-the comparision of LOF, COF and KMEANS algorithms. *Procedia Computer Science*, *176*, 1420-1429.
[6] Machine learning _ Learning notes series (98): Based on connection anomaly factor analysis (Connectivity-Based Outlier Factor) - Liuzhihao (Chih-Hao Liu)
[7] Anomaly detection SOS Algorithm - Hu Guangyue , You know :https://zhuanlan.zhihu.com/p/34438518
[8] Anomaly detection algorithm -- Isolated forests (Isolation Forest) analyse - Risk control big fish , You know :https://zhuanlan.zhihu.com/p/74508141
[9] Isolated forests (isolation Forest)- An algorithm for anomaly detection by randomly dividing a few - Brother Wu talks about risk control , You know :https://zhuanlan.zhihu.com/p/484495545
[10] Isolated forest reading - Mark_Aussie, post :https://blog.csdn.net/MarkAustralia/article/details/12018189
[11] machine learning - Anomaly detection algorithm ( 3、 ... and ):Principal Component Analysis - Liu Tengfei , You know :https://zhuanlan.zhihu.com/p/29091645
[12] Anomaly Detection Anomaly detection --PCA Implementation of algorithm - CC thinking SS, You know :https://zhuanlan.zhihu.com/p/48110105
[13] utilize Autoencoder Perform unsupervised anomaly detection (Python) - SofaSofa.io, You know :https://zhuanlan.zhihu.com/p/46188296
[14] Self encoder AutoEncoder Solve the problem of anomaly detection ( Handlebar handwritten code ) - Data like amber , You know :https://zhuanlan.zhihu.com/p/260882741
[15] Python Machine learning notes :One Class SVM - zoukankan, post :http://t.zoukankan.com/wj-1314-p-10701708.html
[16] Single class SVM: SVDD - Zhang Yice , You know :https://zhuanlan.zhihu.com/p/65617987[17] 【TS Technology class 】 Time series anomaly detection - Timing people , article :https://mp.weixin.qq.com/s/9TimTB_ccPsme2MNPuy6uA
边栏推荐
- 《卓有成效的管理者》读书笔记
- 【无App Push 通用测试方案
- Huawei BFD configuration specification
- Caused by:org.gradle.api.internal.plugins . PluginApplicationException: Failed to apply plugin
- Cognitive introspection
- Function of activation function
- 全链路压测:构建三大模型
- (5) Explanation of yolo-v3 core source code (3)
- 浅谈专项测试之弱网络测试
- The ECU of 21 Audi q5l 45tfsi brushes is upgraded to master special adjustment, and the horsepower is safely and stably increased to 305 horsepower
猜你喜欢
isam2运行流程
MySQL之基础知识
![[ram IP] introduction and experiment of ram IP core](/img/34/1c988456e32a8e9840d1d073caefbf.jpg)
[ram IP] introduction and experiment of ram IP core
误差的基本知识
IP day 16 VLAN MPLS configuration
Embedded point test of app

LAN communication process in the same network segment

The ECU of 21 Audi q5l 45tfsi brushes is upgraded to master special adjustment, and the horsepower is safely and stably increased to 305 horsepower
进程和线程的理解
【微信小程序】搭建开发工具环境
随机推荐
Idea new UI usage
Raised a kitten
MPLS test report
曼哈顿距离和-打印菱形
公司视频加速播放
Eigen sparse matrix operation
Nodejs realizes the third-party login of Weibo
Commodity price visualization
在线问题与离线问题
ContentType的作用
[C language] string left rotation
Title 1093: character reverse order
Configuring OSPF GR features for Huawei devices
LAN communication process in the same network segment
leaflet 地图
Detailed explanation of BF and KMP
技术分享 | 常见接口协议解析
Web界面元素的测试
[wechat applet] build a development tool environment
【C语言】字符串左旋