author |THILAKADIBOINA
compile |Flin
source |analyticsvidhya
Introduce
This paper introduces the generative countermeasure network (Generative attersarial Networks,GAN) Use , This is a kind of real Covid-19 The technique of oversampling data , Used to predict mortality . This story gives us a better understanding of the data preparation steps ( Such as dealing with unbalanced data ) How to improve model performance .
The data and core model of this article are from Celestine Iwendi、Ali Kashif Bashir、Atharva Peshkar A recent study (2020 year 7 month )“ Using enhanced random forest algorithm to predict COVID-19 Healthy patients ”. Used in this study ADABOST The model enhanced random forest algorithm predicts the mortality of individual patients , Accuracy rate is 94%. This paper considers the same model and model parameters , Based on the analysis of the use of GAN The improvement of the existing model by oversampling technology of .
For aspiring data scientists , One of the best ways to learn good practice is to participate in hacker competitions on different forums , such as Vidhya、Kaggle Or other forums .
Besides , Obtain resolved cases and data from these forums or published research publications ; How to get to know them , And try to improve the accuracy or reduce the error through additional steps . This will form a solid foundation , It enables us to think deeply about the application of other technologies we have learned in the value chain of Data Science .
The data used in the study was 222 Of a patient 13 To train with . The data is biased ,159 example (72%) Belong to “0” Class or “ Has resumed ” class . Because of its deviant nature , All kinds of undersampling / Oversampling can be applied to data . The problem of skewed data will lead to over fitting of prediction model .
To overcome this limitation , Many studies use oversampling to balance data sets , In order to get more accurate model training . Oversampling is a technique that compensates for data set imbalance by increasing the number of samples in a small number of data .
Conventional methods include random oversampling (ROS)、 Synthesis of a few oversampling techniques (SMOTE) etc. . For more information about handling unbalanced classes using conventional methods , see also :
lately , A machine learning model of generative network based on antagonistic learning concept is proposed , It's a generative adversarial network . Generative countermeasures network (Generative atterial Networks,GAN) It is easy to apply to oversampling research , Because the nature of the neural network based on confrontation training allows the generation of artificial data similar to the original data . The oversampling based on generative countermeasure network overcomes the traditional method ( If over fitting ) The limitations of , It allows the establishment of a high-precision prediction model for unbalanced data .
How to generate composite data ?
Two neural networks compete with each other , Learning goal distribution and generating artificial data
Generator network G: Simulation training sample deception discriminator
Discrimination network D: Distinguish training samples from generated samples
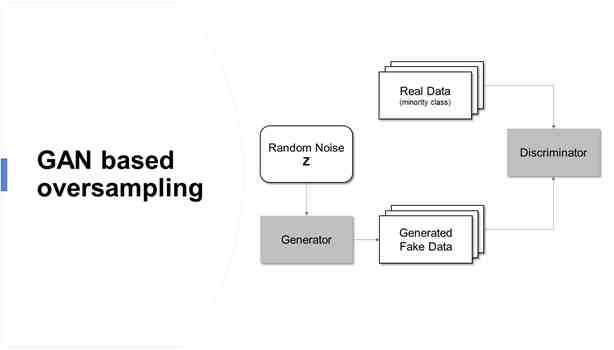
Generative antagonistic networks are scenarios based on game theory , Among them, the generating network must compete with its competitors . With GAN Learn to simulate the distribution of data , It's used in all kinds of fields , Such as music 、 Video and natural language , Recently, it has also been used to deal with unbalanced data problems .
The data and basic models used in the study can be found here
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow as tf
from keras.layers import Input, Dense, Reshape, Flatten, Dropout, BatchNormalization, Embedding
from keras.layers.advanced_activations import LeakyReLU
from keras.layers.merge import concatenate
from keras.models import Sequential, Model
from keras.optimizers import Adam
from keras.utils import to_categorical
from keras.layers.advanced_activations import LeakyReLU
from keras.utils.vis_utils import plot_model
from sklearn.preprocessing import MinMaxScaler, OneHotEncoder, LabelEncoder
import scipy.stats
import datetime as dt
import pydot
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
df = pd.read_csv('Covid_Train_Oct32020.csv')
df = df.drop('id',axis=1)
df = df.fillna(np.nan,axis=0)
df['age'] = df['age'].fillna(value=df['age'].mean())
df['sym_on'] = pd.to_datetime(df['sym_on'])
df['hosp_vis'] = pd.to_datetime(df['hosp_vis'])
df['sym_on']= df['sym_on'].map(dt.datetime.toordinal)
df['hosp_vis']= df['hosp_vis'].map(dt.datetime.toordinal)
df['diff_sym_hos']= df['hosp_vis'] - df['sym_on']
df=df.drop(['sym_on', 'hosp_vis'], axis=1)
df['location'] = df['location'].astype(str)
df['country'] = df['country'].astype(str)
df['gender'] = df['gender'].astype(str)
df['vis_wuhan'] = df['vis_wuhan'].astype(str)
df['from_wuhan'] = df['from_wuhan'].astype(str)
df['symptom1'] = df['symptom1'].astype(str)
df['symptom2'] = df['symptom2'].astype(str)
df['symptom3'] = df['symptom3'].astype(str)
df['symptom4'] = df['symptom4'].astype(str)
df['symptom5'] = df['symptom5'].astype(str)
df['symptom6'] = df['symptom6'].astype(str)
df.dtypesData description
| Column | describe | value ( Used to categorize variables ) | type |
| id | Patient number | Do not apply | Numbers |
| location | Where the patient belongs | Cities all over the world | character string , classification |
| country | The country of the patient | Several countries | character string , classification |
| gender | The patient's gender | male , Woman | character string , classification |
| age | Patient age | Do not apply | Numbers |
| sym_on | The date the patient began to notice the symptoms | Do not apply | date |
| hosp_vis | The date the patient went to the hospital | Do not apply | date |
| vis_wuhan | Has the patient ever been to Wuhan, China | yes (1), no (0) | The number , classification |
| from_wuhan | Whether the patient belongs to Wuhan, China | yes (1), no (0) | The number , classification |
| death | Is the patient due to COVID-19 And die | yes (1), no (0) | The number , classification |
| Recov | Whether the patient has recovered | yes (1), no (0) | The number , classification |
| symptom1. symptom2, symptom3, symptom4, symptom5, symptom6 | Symptoms noticed by the patient | Patients notice a variety of symptoms | character string , classification |
The study considered 11 Classification input features and 2 Digital input features . The target variable is death / recovery . New column filled “ diff_sym_hos”, To provide the difference between the symptoms found and received in the hospital that day .
The focus of the study is to improve a few categories of data , Death == 1, A subset is extracted from the training data . Subsets are separated by category and number , And pass it on to GAN Model .
df_minority_data=df.loc[df['death'] == 1]
#Subsetting input features without target variable
df_minority_data_withouttv=df_minority_data.loc[:, df_minority_data.columns != 'death']
numerical_df = df_minority_data_withouttv.select_dtypes("number")
categorical_df = df_minority_data_withouttv.select_dtypes("object")
scaling = MinMaxScaler()
numerical_df_rescaled = scaling.fit_transform(numerical_df)
get_dummy_df = pd.get_dummies(categorical_df)
#Seperating Each Category
location_dummy_col = [col for col in get_dummy_df.columns if 'location' in col]
location_dummy = get_dummy_df[location_dummy_col]
country_dummy_col = [col for col in get_dummy_df.columns if 'country' in col]
country_dummy = get_dummy_df[country_dummy_col]
gender_dummy_col = [col for col in get_dummy_df.columns if 'gender' in col]
gender_dummy = get_dummy_df[gender_dummy_col]
vis_wuhan_dummy_col = [col for col in get_dummy_df.columns if 'vis_wuhan' in col]
vis_wuhan_dummy = get_dummy_df[vis_wuhan_dummy_col]
from_wuhan_dummy_col = [col for col in get_dummy_df.columns if 'from_wuhan' in col]
from_wuhan_dummy = get_dummy_df[from_wuhan_dummy_col]
symptom1_dummy_col = [col for col in get_dummy_df.columns if 'symptom1' in col]
symptom1_dummy = get_dummy_df[symptom1_dummy_col]
symptom2_dummy_col = [col for col in get_dummy_df.columns if 'symptom2' in col]
symptom2_dummy = get_dummy_df[symptom2_dummy_col]
symptom3_dummy_col = [col for col in get_dummy_df.columns if 'symptom3' in col]
symptom3_dummy = get_dummy_df[symptom3_dummy_col]
symptom4_dummy_col = [col for col in get_dummy_df.columns if 'symptom4' in col]
symptom4_dummy = get_dummy_df[symptom4_dummy_col]
symptom5_dummy_col = [col for col in get_dummy_df.columns if 'symptom5' in col]
symptom5_dummy = get_dummy_df[symptom5_dummy_col]
symptom6_dummy_col = [col for col in get_dummy_df.columns if 'symptom6' in col]
symptom6_dummy = get_dummy_df[symptom6_dummy_col]
Define generator
The generator takes input from potential space and generates a new composite sample . Leak correction linear element (LeakyReLU) It's a function used in generator and discriminator models to deal with some negative values .
It uses the default recommended value 0.2 And the appropriate weight initialization procedure “ he_uniform” Use . Besides , Use batch normalization between layers to standardize activation from previous layers ( Zero mean and unit variance ) And stabilize the training process .
In the output layer ,softmax Activation functions are used to categorize variables , and sigmoid Functions are used for continuous variables .
def define_generator (catsh1,catsh2,catsh3,catsh4,catsh5,catsh6,catsh7,catsh8,catsh9,catsh10,catsh11,numerical):
#Inputting noise from latent space
noise = Input(shape = (70,))
hidden_1 = Dense(8, kernel_initializer = "he_uniform")(noise)
hidden_1 = LeakyReLU(0.2)(hidden_1)
hidden_1 = BatchNormalization(momentum = 0.8)(hidden_1)
hidden_2 = Dense(16, kernel_initializer = "he_uniform")(hidden_1)
hidden_2 = LeakyReLU(0.2)(hidden_2)
hidden_2 = BatchNormalization(momentum = 0.8)(hidden_2)
#Branch 1 for generating location data
branch_1 = Dense(32, kernel_initializer = "he_uniform")(hidden_2)
branch_1 = LeakyReLU(0.2)(branch_1)
branch_1 = BatchNormalization(momentum = 0.8)(branch_1)
branch_1 = Dense(64, kernel_initializer = "he_uniform")(branch_1)
branch_1 = LeakyReLU(0.2)(branch_1)
branch_1 = BatchNormalization(momentum=0.8)(branch_1)
#Output Layer1
branch_1_output = Dense(catsh1, activation = "softmax")(branch_1)
#Likewise, for all remaining 10 categories branches will be defined
#Branch 12 for generating numerical data
branch_12 = Dense(64, kernel_initializer = "he_uniform")(hidden_2)
branch_12 = LeakyReLU(0.2)(branch_3)
branch_12 = BatchNormalization(momentum=0.8)(branch_12)
branch_12 = Dense(128, kernel_initializer = "he_uniform")(branch_12)
branch_12 = LeakyReLU(0.2)(branch_12)
branch_12 = BatchNormalization(momentum=0.8)(branch_12)
#Output Layer12
branch_12_output = Dense(numerical, activation = "sigmoid")(branch_12)
#Combined output
combined_output = concatenate([branch_1_output, branch_2_output, branch_3_output,branch_4_output,branch_5_output,branch_6_output,branch_7_output,branch_8_output,branch_9_output,branch_10_output,branch_11_output,branch_12_output])
#Return model
return Model(inputs = noise, outputs = combined_output)
generator = define_generator(location_dummy.shape[1],country_dummy.shape[1],gender_dummy.shape[1],vis_wuhan_dummy.shape[1],from_wuhan_dummy.shape[1],symptom1_dummy.shape[1],symptom2_dummy.shape[1],symptom3_dummy.shape[1],symptom4_dummy.shape[1],symptom5_dummy.shape[1],symptom6_dummy.shape[1],numerical_df_rescaled.shape[1])
generator.summary()Defining discriminators
The discriminator model will be derived from our data ( For example, vector ) Get samples from , And output the classification prediction about whether the sample is true or false . It's a binary classification problem , So use... In the output layer sigmoid Activation function , The binary cross entropy loss function is used in the model compilation . Use the learning rate LR by 0.0002 And suggested beta1 The momentum is 0.5 Of Adam optimization algorithm .
def define_discriminator(inputs_n):
#Input from generator
d_input = Input(shape = (inputs_n,))
d = Dense(128, kernel_initializer="he_uniform")(d_input)
d = LeakyReLU(0.2)(d)
d = Dense(64, kernel_initializer="he_uniform")(d)
d = LeakyReLU(0.2)(d)
d = Dense(32, kernel_initializer="he_uniform")(d)
d = LeakyReLU(0.2)(d)
d = Dense(16, kernel_initializer="he_uniform")(d)
d = LeakyReLU(0.2)(d)
d = Dense(8, kernel_initializer="he_uniform")(d)
d = LeakyReLU(0.2)(d)
#Output Layer
d_output = Dense(1, activation = "sigmoid")(d)
#compile and return model
model = Model(inputs = d_input, outputs = d_output)
model.compile(loss = "binary_crossentropy", optimizer = Adam(lr=0.0002, beta_1=0.5), metrics = ["accuracy"])
return model
inputs_n = location_dummy.shape[1]+country_dummy.shape[1]+gender_dummy.shape[1]+vis_wuhan_dummy.shape[1]+from_wuhan_dummy.shape[1]+symptom1_dummy.shape[1]+symptom2_dummy.shape[1]+symptom3_dummy.shape[1]+symptom4_dummy.shape[1]+symptom5_dummy.shape[1]+symptom6_dummy.shape[1]+numerical_df_rescaled.shape[1]
discriminator = define_discriminator(inputs_n)
discriminator.summary()Combine the generator and discriminator into GAN Model and complete the training . Considering 7,000 Period , And consider the complete minority training data .
Def define_complete_gan(generator, discriminator):
discriminator.trainable = False
gan_output = discriminator(generator.output)
#Initialize gan
model = Model(inputs = generator.input, outputs = gan_output)
#Model Compilation
model.compile(loss = "binary_crossentropy", optimizer = Adam(lr=0.0002, beta_1=0.5))
return model
completegan = define_complete_gan(generator, discriminator)
def gan_train(gan, generator, discriminator, catsh1,catsh2,catsh3,catsh4,catsh5,catsh6,catsh7,catsh8,catsh9,catsh10,catsh11,numerical, latent_dim, n_epochs, n_batch, n_eval):
#Upddte Discriminator with half batch size
half_batch = int(n_batch / 2)
discriminator_loss = []
generator_loss = []
#generate class labels for fake and real
valid = np.ones((half_batch, 1))
y_gan = np.ones((n_batch, 1))
fake = np.zeros((half_batch, 1))
#training
for i in range(n_epochs):
#select random batch from real categorical and numerical data
idx = np.random.randint(0, catsh1.shape[0], half_batch)
location_real = catsh1[idx]
country_real = catsh2[idx]
gender_real = catsh3[idx]
vis_wuhan_real = catsh4[idx]
from_wuhan_real = catsh5[idx]
symptom1_real = catsh6[idx]
symptom2_real = catsh7[idx]
symptom3_real = catsh8[idx]
symptom4_real = catsh9[idx]
symptom5_real = catsh10[idx]
symptom6_real = catsh11[idx]
numerical_real = numerical_df_rescaled[idx]
#concatenate categorical and numerical data for the discriminator
real_data = np.concatenate([location_real, country_real, gender_real,vis_wuhan_real,from_wuhan_real,symptom1_real,symptom2_real,symptom3_real,symptom4_real,symptom5_real,symptom6_real,numerical_real], axis = 1)
#generate fake samples from the noise
noise = np.random.normal(0, 1, (half_batch, latent_dim))
fake_data = generator.predict(noise)
#train the discriminator and return losses and acc
d_loss_real, da_real = discriminator.train_on_batch(real_data, valid)
d_loss_fake, da_fake = discriminator.train_on_batch(fake_data, fake)
d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)
discriminator_loss.append(d_loss)
#generate noise for generator input and train the generator (to have the discriminator label samples as valid)
noise = np.random.normal(0, 1, (n_batch, latent_dim))
g_loss = gan.train_on_batch(noise, y_gan)
generator_loss.append(g_loss)
#evaluate progress
if (i+1) % n_eval == 0:
print ("Epoch: %d [Discriminator loss: %f] [Generator loss: %f]" % (i + 1, d_loss, g_loss))
plt.figure(figsize = (20, 10))
plt.plot(generator_loss, label = "Generator loss")
plt.plot(discriminator_loss, label = "Discriminator loss")
plt.title("Stats from training GAN")
plt.grid()
plt.legend()
latent_dim = 100
gan_train(completegan, generator, discriminator, location_dummy.values,country_dummy.values,gender_dummy.values,vis_wuhan_dummy.values,from_wuhan_dummy.values,symptom1_dummy.values,symptom2_dummy.values,symptom3_dummy.values,symptom4_dummy.values,symptom5_dummy.values,symptom6_dummy.values,numerical_df_rescaled, latent_dim, n_epochs = 7000, n_batch = 63, n_eval = 200)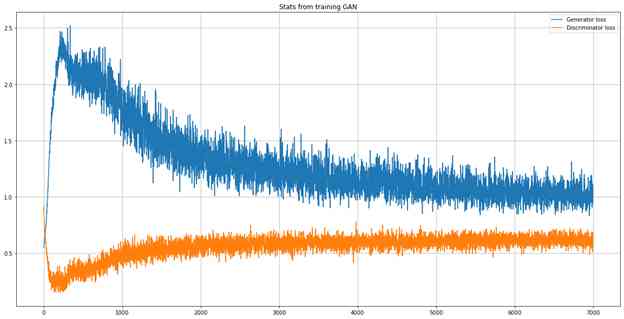
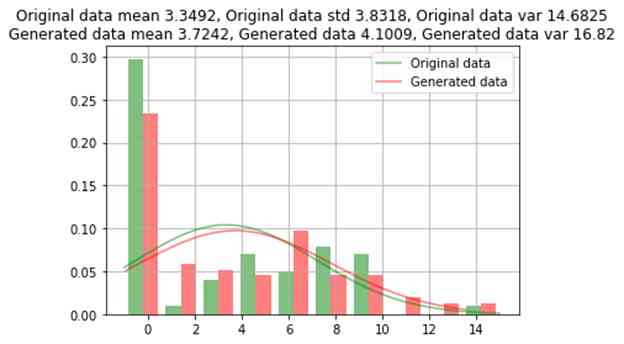
The trained model is used to generate a few other 96 Bar record , To divide each class equally (159). Now the generated numerical data and the mean value of the original data , Compare the standard deviation with the variance ; And compare the category data according to the count of each category .
noise = np.random.normal(0, 1, (96, 100))
generated_mixed_data = generator.predict(noise)
columns=list(location_dummy.columns)+list(country_dummy.columns)+list(gender_dummy.columns)+list(vis_wuhan_dummy.columns)+list(from_wuhan_dummy.columns)+list(symptom1_dummy.columns)+list(symptom2_dummy.columns)+list(symptom3_dummy.columns)+list(symptom4_dummy.columns)+list(symptom5_dummy.columns)+list(symptom6_dummy.columns)+list(numerical_df.columns)
mixed_gen_df = pd.DataFrame(data = generated_mixed_data, columns = columns)
mixed_gen_df.iloc[:,:-3] = np.round(mixed_gen_df.iloc[:,:-3])
mixed_gen_df.iloc[:,-2:] = scaling.inverse_transform(mixed_gen_df.iloc[:,-2:])
#Original Data
original_df = pd.concat([location_dummy,country_dummy,gender_dummy,vis_wuhan_dummy,from_wuhan_dummy,symptom1_dummy,symptom2_dummy,symptom3_dummy,symptom4_dummy,symptom5_dummy,symptom6_dummy,numerical_df], axis = 1)
def normal_distribution(org, noise):
org_x = np.linspace(org.min(), org.max(), len(org))
noise_x = np.linspace(noise.min(), noise.max(), len(noise))
org_y = scipy.stats.norm.pdf(org_x, org.mean(), org.std())
noise_y = scipy.stats.norm.pdf(noise_x, noise.mean(), noise.std())
n, bins, patches = plt.hist([org, noise], density = True, alpha = 0.5, color = ["green", "red"])
xmin, xmax = plt.xlim()
plt.plot(org_x, org_y, color = "green", label = "Original data", alpha = 0.5)
plt.plot(noise_x, noise_y, color = "red", label = "Generated data", alpha = 0.5)
title = f"Original data mean {np.round(org.mean(), 4)}, Original data std {np.round(org.std(), 4)}, Original data var {np.round(org.var(), 4)}\nGenerated data mean {np.round(noise.mean(), 4)}, Generated data {np.round(noise.std(), 4)}, Generated data var {np.round(noise.var(), 2)}"
plt.title(title)
plt.legend()
plt.grid()
plt.show()
Numeric_columns=numerical_df.columns
for column in numerical_df.columns:
print(column, "Comparison between Original Data and Generated Data")
normal_distribution(original_df
, mixed_gen_df
)Age comparison between raw data and generated data
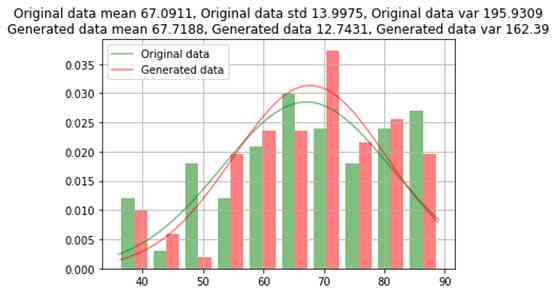
Comparison between raw data and generated data
Category comparison between raw data and generated data
| features | Raw data | Data generated | ||
| 0 | 1 | 0 | 1 | |
| location_Hokkaido | 61 | 2 | 95 | 1 |
| gender_female | 49 | 14 | 60 | 36 |
| symptom2_ cough | 62 | 1 | 96 | 0 |
GAN The data generated by the oversampling method is almost similar to the original data , The error of the original data is about 1%. For some rare categories , Data will not be generated on all category values .
Follow the same data preparation steps as mentioned in the original study , To see through the use of GAN How does supersampling improve the performance of the model compared with the original method . The unique hot coded data of the generated sample is converted to the original data frame format .
# Getting Back Categorical Data in Original_Format from Dummies
location_filter_col = [col for col in mixed_gen_df if col.startswith('location')]
location=mixed_gen_df[location_filter_col]
location= pd.get_dummies(location).idxmax(1)
location= location.replace('location_', '', regex=True)
df_generated_data = pd.DataFrame()
df_generated_data['location']=location
country_filter_col = [col for col in mixed_gen_df if col.startswith('country')]
country=mixed_gen_df[country_filter_col]
country= pd.get_dummies(country).idxmax(1)
country= country.replace('country_', '', regex=True)
df_generated_data['country']=country
gender_filter_col = [col for col in mixed_gen_df if col.startswith('gender')]
gender=mixed_gen_df[gender_filter_col]
gender= pd.get_dummies(gender).idxmax(1)
gender= gender.replace('gender_', '', regex=True)
df_generated_data['gender']=gender
vis_wuhan_filter_col = [col for col in mixed_gen_df if col.startswith('vis_wuhan')]
vis_wuhan=mixed_gen_df[vis_wuhan_filter_col]
vis_wuhan= pd.get_dummies(vis_wuhan).idxmax(1)
vis_wuhan= vis_wuhan.replace('vis_wuhan_', '', regex=True)
df_generated_data['vis_wuhan']=vis_wuhan
from_wuhan_filter_col = [col for col in mixed_gen_df if col.startswith('from_wuhan')]
from_wuhan=mixed_gen_df[from_wuhan_filter_col]
from_wuhan= pd.get_dummies(from_wuhan).idxmax(1)
from_wuhan= from_wuhan.replace('from_wuhan_', '', regex=True)
df_generated_data['from_wuhan']=from_wuhan
symptom1_filter_col = [col for col in mixed_gen_df if col.startswith('symptom1')]
symptom1=mixed_gen_df[symptom1_filter_col]
symptom1= pd.get_dummies(symptom1).idxmax(1)
symptom1= symptom1.replace('symptom1_', '', regex=True)
df_generated_data['symptom1']=symptom1
symptom2_filter_col = [col for col in mixed_gen_df if col.startswith('symptom2')]
symptom2=mixed_gen_df[symptom2_filter_col]
symptom2= pd.get_dummies(symptom2).idxmax(1)
symptom2= symptom2.replace('symptom2_', '', regex=True)
df_generated_data['symptom2']=symptom2
symptom3_filter_col = [col for col in mixed_gen_df if col.startswith('symptom3')]
symptom3=mixed_gen_df[symptom3_filter_col]
symptom3= pd.get_dummies(symptom3).idxmax(1)
symptom3= symptom3.replace('symptom3_', '', regex=True)
df_generated_data['symptom3']=symptom3
symptom4_filter_col = [col for col in mixed_gen_df if col.startswith('symptom4')]
symptom4=mixed_gen_df[symptom4_filter_col]
symptom4= pd.get_dummies(symptom4).idxmax(1)
symptom4= symptom4.replace('symptom4_', '', regex=True)
df_generated_data['symptom4']=symptom4
symptom5_filter_col = [col for col in mixed_gen_df if col.startswith('symptom5')]
symptom5=mixed_gen_df[symptom5_filter_col]
symptom5= pd.get_dummies(symptom5).idxmax(1)
symptom5= symptom5.replace('symptom5_', '', regex=True)
df_generated_data['symptom5']=symptom5
symptom6_filter_col = [col for col in mixed_gen_df if col.startswith('symptom6')]
symptom6=mixed_gen_df[symptom6_filter_col]
symptom6= pd.get_dummies(symptom6).idxmax(1)
symptom6= symptom6.replace('symptom6_', '', regex=True)
df_generated_data['symptom6']=symptom6
df_generated_data['death']=1
df_generated_data['death']=1
df_generated_data[['age','diff_sym_hos']]=mixed_gen_df[['age','diff_sym_hos']]
df_generated_data = df_generated_data.fillna(np.nan,axis=0)
#Encoding Data
encoder_location = preprocessing.LabelEncoder()
encoder_country = preprocessing.LabelEncoder()
encoder_gender = preprocessing.LabelEncoder()
encoder_symptom1 = preprocessing.LabelEncoder()
encoder_symptom2 = preprocessing.LabelEncoder()
encoder_symptom3 = preprocessing.LabelEncoder()
encoder_symptom4 = preprocessing.LabelEncoder()
encoder_symptom5 = preprocessing.LabelEncoder()
encoder_symptom6 = preprocessing.LabelEncoder()
# Loading and Preparing Data
df = pd.read_csv('Covid_Train_Oct32020.csv')
df = df.drop('id',axis=1)
df = df.fillna(np.nan,axis=0)
df['age'] = df['age'].fillna(value=tdata['age'].mean())
df['sym_on'] = pd.to_datetime(df['sym_on'])
df['hosp_vis'] = pd.to_datetime(df['hosp_vis'])
df['sym_on']= df['sym_on'].map(dt.datetime.toordinal)
df['hosp_vis']= df['hosp_vis'].map(dt.datetime.toordinal)
df['diff_sym_hos']= df['hosp_vis'] - df['sym_on']
df = df.drop(['sym_on','hosp_vis'],axis=1)
df['location'] = encoder_location.fit_transform(df['location'].astype(str))
df['country'] = encoder_country.fit_transform(df['country'].astype(str))
df['gender'] = encoder_gender.fit_transform(df['gender'].astype(str))
df[['symptom1']] = encoder_symptom1.fit_transform(df['symptom1'].astype(str))
df[['symptom2']] = encoder_symptom2.fit_transform(df['symptom2'].astype(str))
df[['symptom3']] = encoder_symptom3.fit_transform(df['symptom3'].astype(str))
df[['symptom4']] = encoder_symptom4.fit_transform(df['symptom4'].astype(str))
df[['symptom5']] = encoder_symptom5.fit_transform(df['symptom5'].astype(str))
df[['symptom6']] = encoder_symptom6.fit_transform(df['symptom6'].astype(str))
# Encoding Generated Data
df_generated_data['location'] = encoder_location.transform(df_generated_data['location'].astype(str))
df_generated_data['country'] = encoder_country.transform(df_generated_data['country'].astype(str))
df_generated_data['gender'] = encoder_gender.transform(df_generated_data['gender'].astype(str))
df_generated_data[['symptom1']] = encoder_symptom1.transform(df_generated_data['symptom1'].astype(str))
df_generated_data[['symptom2']] = encoder_symptom2.transform(df_generated_data['symptom2'].astype(str))
df_generated_data[['symptom3']] = encoder_symptom3.transform(df_generated_data['symptom3'].astype(str))
df_generated_data[['symptom4']] = encoder_symptom4.transform(df_generated_data['symptom4'].astype(str))
df_generated_data[['symptom5']] = encoder_symptom5.transform(df_generated_data['symptom5'].astype(str))
df_generated_data[['symptom6']] = encoder_symptom6.transform(df_generated_data['symptom6'].astype(str))
df_generated_data[['diff_sym_hos']] = df_generated_data['diff_sym_hos'].astype(int)Model comparison
Divide the raw data into training and testing , take GAN The generated data is added to the training data , To compare performance to the basic model . In practice ( original ) Test model performance on split test data .
from sklearn.metrics import recall_score as rs
from sklearn.metrics import precision_score as ps
from sklearn.metrics import f1_score as fs
from sklearn.metrics import balanced_accuracy_score as bas
from sklearn.metrics import confusion_matrix as cm
import numpy as np
import pandas as pd
import datetime as dt
import sklearn
from scipy import stats
from sklearn import preprocessing
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import recall_score as rs
from sklearn.metrics import precision_score as ps
from sklearn.metrics import f1_score as fs
from sklearn.metrics import log_loss
rf = RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=2, max_features='auto',
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=2, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=100,
n_jobs=None, oob_score=False, random_state=None,
verbose=0, warm_start=False)
classifier = AdaBoostClassifier(rf,50,0.01,'SAMME.R',10)
#Seperate TV in Generated Data
X1 = df_generated_data.loc[:, df_generated_data.columns != 'death']
Y1 = df_generated_data['death']
#Seperate TV in Original Data
X = df.loc[:, df.columns != 'death']
Y = df['death']
#Splitting Original Data
X_train, X_test, Y_train, Y_test = train_test_split(X,Y,test_size=0.2,random_state=0)
#Appending Generated Data to X_train
X_train1=X_train.append(X1, sort=False)
Y_train1=Y_train.append(Y1)
classifier.fit(X_train1,np.array(Y_train1).reshape(Y_train1.shape[0],1))
pred = np.array(classifier.predict(X_test))
recall = rs(Y_test,pred)
precision = ps(Y_test,pred)
r1 = fs(Y_test,pred)
ma = classifier.score(X_test,Y_test)
print('*** Evaluation metrics for test dataset ***\n')
print('Recall Score: ',recall)
print('Precision Score: ',precision)
print('F1 Score: ',f1)
print('Accuracy: ',ma)| Metric system | Basic model score * | Scoring with enhanced generated data |
| Recall score | 0.75 | 0.83 |
| Precision fraction | 1 | 1 |
| F1 fraction | 0.86 | 0.9 |
| accuracy | 0.9 | 0.95 |
source : surface 3 Basic model indicators
Conclusion
Compared with the basic model , The proposed model provides more accurate and reliable results , Show based on GAN Over sampling overcomes the limitation of unbalanced data , And appropriately extend a few classes .
Link to the original text :https://www.analyticsvidhya.c...
Welcome to join us AI Blog station :
http://panchuang.net/
sklearn Machine learning Chinese official documents :
http://sklearn123.com/
Welcome to pay attention to pan Chuang blog resource summary station :
http://docs.panchuang.net/