当前位置:网站首页>[paper reading] cavemix: a simple data augmentation method for brain vision segmentation
[paper reading] cavemix: a simple data augmentation method for brain vision segmentation
2022-07-04 23:54:00 【xiongxyowo】
[ Address of thesis ][ Code ][MICCAI 21]
Abstract
Segmentation of brain lesions (Brain Lesion Segmentation) It provides a valuable tool for clinical diagnosis , Convolutional neural networks (CNN) We have achieved unprecedented success in this task . Data enhancement is a widely used strategy , Can improve CNN Training effect , The design of enhancement methods for brain lesion segmentation is still an open problem . In this work , We propose a simple data enhancement method , go by the name of CarveMix, Used based on CNN Segmentation of brain lesions . With others based on " blend " The same way , Such as Mixup and CutMix,CarveMix Randomly combine two existing marker images to generate new marker samples . However , Different from these image combination based enhancement strategies ,CarveMix It is disease perception , Pay attention to pathological changes when combining , And create appropriate annotations for the generated image . say concretely , According to the location and geometry of the lesion , Carve a region of interest from a marked image (ROI),ROI The size of is sampled from a probability distribution . then , Carved ROI It replaces the corresponding voxel in the second marked image , The annotation of the second image is also replaced accordingly . In this way , We generate new tagged images for network training , And the lesion information is preserved . In order to evaluate the proposed method , We conducted experiments on two brain lesion datasets . It turns out that , Compared with other simple data enhancement methods , Our method improves the accuracy of segmentation .
Method
This paper is a data enhancement method specially proposed for brain lesion segmentation ——CarveMix. This method is also a method based on label fusion , such as MixUp(ICLR 18) Is to fuse the two labels linearly , and CarveMix(ICCV 19) It is a kind of nonlinear fusion . It should be noted that , These classical methods are used for image classification tasks , Therefore, there is a lack of label fusion methods for segmentation tasks .CarveMix The integration process of is as follows :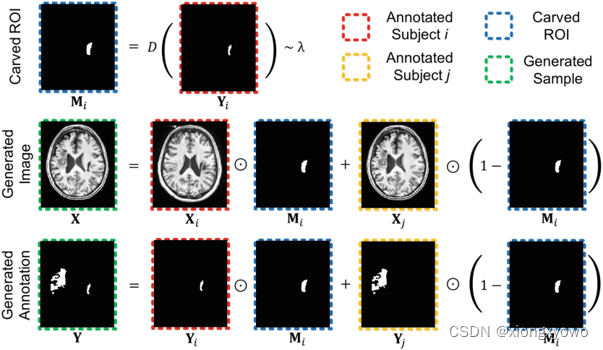
First look at the formula directly : Fused image X \mathbf{X} X And the label obtained by fusion Y \mathbf{Y} Y The final calculation process of is as follows : X = X i ⊙ M i + X j ⊙ ( 1 − M i ) \mathbf{X}=\mathbf{X}_{i} \odot \mathbf{M}_{i}+\mathbf{X}_{j} \odot\left(1-\mathbf{M}_{i}\right) X=Xi⊙Mi+Xj⊙(1−Mi) Y = Y i ⊙ M i + Y j ⊙ ( 1 − M i ) \mathbf{Y}=\mathbf{Y}_{i} \odot \mathbf{M}_{i}+\mathbf{Y}_{j} \odot\left(1-\mathbf{M}_{i}\right) Y=Yi⊙Mi+Yj⊙(1−Mi) For the sake of intuition , We take label fusion as an example to show the specific fusion process , The fusion of image itself is consistent with the fusion of label . Seen from the figure , The fusion of labels is basically equivalent to directly merging two original images mask Y i \mathbf{Y}_i Yi and Y j \mathbf{Y}_j Yj Add it up directly :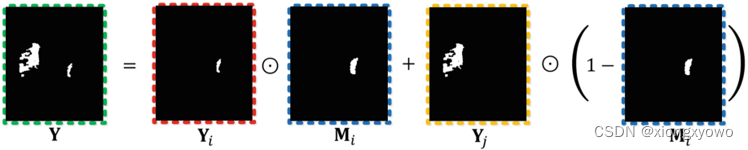
In fact, it is Y i \mathbf{Y}_i Yi Multiplied by a factor a a a After and Y j \mathbf{Y}_j Yj Multiplied by a factor b b b And then add up , Yes a + b = 1 a+b=1 a+b=1. In the figure "⊙" The symbol represents pixel by pixel , So you can even put M i \mathbf{M}_i Mi As a spatial attention map .
The problem now is actually how to calculate M i \mathbf{M}_i Mi 了 . As you can see from the diagram , M i \mathbf{M}_i Mi And Y i \mathbf{Y}_i Yi In fact, it is very similar , It's a bit like in M i \mathbf{M}_i Mi An expansion operation is carried out on the basis of . say concretely , M i \mathbf{M}_i Mi pass the civil examinations j j j A pixel value M i v \mathbf{M}_i^v Miv The calculation method is as follows : M i v = { 1 , D v ( Y i ) ≤ λ 0 , otherwise \mathbf{M}_{i}^{v}=\left\{\begin{array}{l} 1, D^{v}\left(\mathbf{Y}_{i}\right) \leq \lambda \\ 0, \text { otherwise } \end{array}\right. Miv={ 1,Dv(Yi)≤λ0, otherwise D v ( Y i ) = { − d ( v , ∂ Y i ) , if Y i v = 1 d ( v , ∂ Y i ) , if Y i v = 0 D^{v}\left(\mathbf{Y}_{i}\right)=\left\{\begin{aligned} -d\left(v, \partial \mathbf{Y}_{i}\right), & \text { if } \mathbf{Y}_{i}^{v}=1 \\ d\left(v, \partial \mathbf{Y}_{i}\right), & \text { if } \mathbf{Y}_{i}^{v}=0 \end{aligned}\right. Dv(Yi)={ −d(v,∂Yi),d(v,∂Yi), if Yiv=1 if Yiv=0 This d ( v , ∂ Y i ) d(v, \partial \mathbf{Y}_{i}) d(v,∂Yi) Refers to the current pixel v v v And " Lesion boundary " ∂ Y i \partial \mathbf{Y}_{i} ∂Yi Distance of . You can see , If v v v Itself is in the lesion area , that D v ( Y i ) D^{v}\left(\mathbf{Y}_{i}\right) Dv(Yi) Just give a negative number , So as to ensure that it has a greater probability of being selected ( That is less than λ \lambda λ); And if it is outside the lesion area , We think that the closer we are, the more we should be selected . It is worth noting that , This λ \lambda λ It's positive and negative , So as to ensure the pathological area " inflation " perhaps " shrinkage ". λ \lambda λ The specific calculation process of is more complex , Interested readers can read the original .
边栏推荐
- 多回路仪表在基站“转改直”方面的应用
- [kotlin] the third day
- Blue sky nh55 series notebook memory reading and writing speed is extremely slow, solution process record
- Advanced template
- MariaDB's Galera cluster application scenario -- multi master and multi active databases
- How long does it take to obtain a PMP certificate?
- Pict generate orthogonal test cases tutorial
- 公司要上监控,Zabbix 和 Prometheus 怎么选?这么选准没错!
- 【雅思阅读】王希伟阅读P4(matching2段落信息配对题【困难】)
- Compare two vis in LabVIEW
猜你喜欢

Why does infographic help your SEO
电力运维云平台:开启电力系统“无人值班、少人值守”新模式

Combien de temps faut - il pour obtenir un certificat PMP?
高配笔记本使用CAD搬砖时卡死解决记录
业务场景功能的继续修改

企业公司项目开发好一部分基础功能,重要的事保存到线上第一a

Application of fire fighting system based on 3D GIS platform
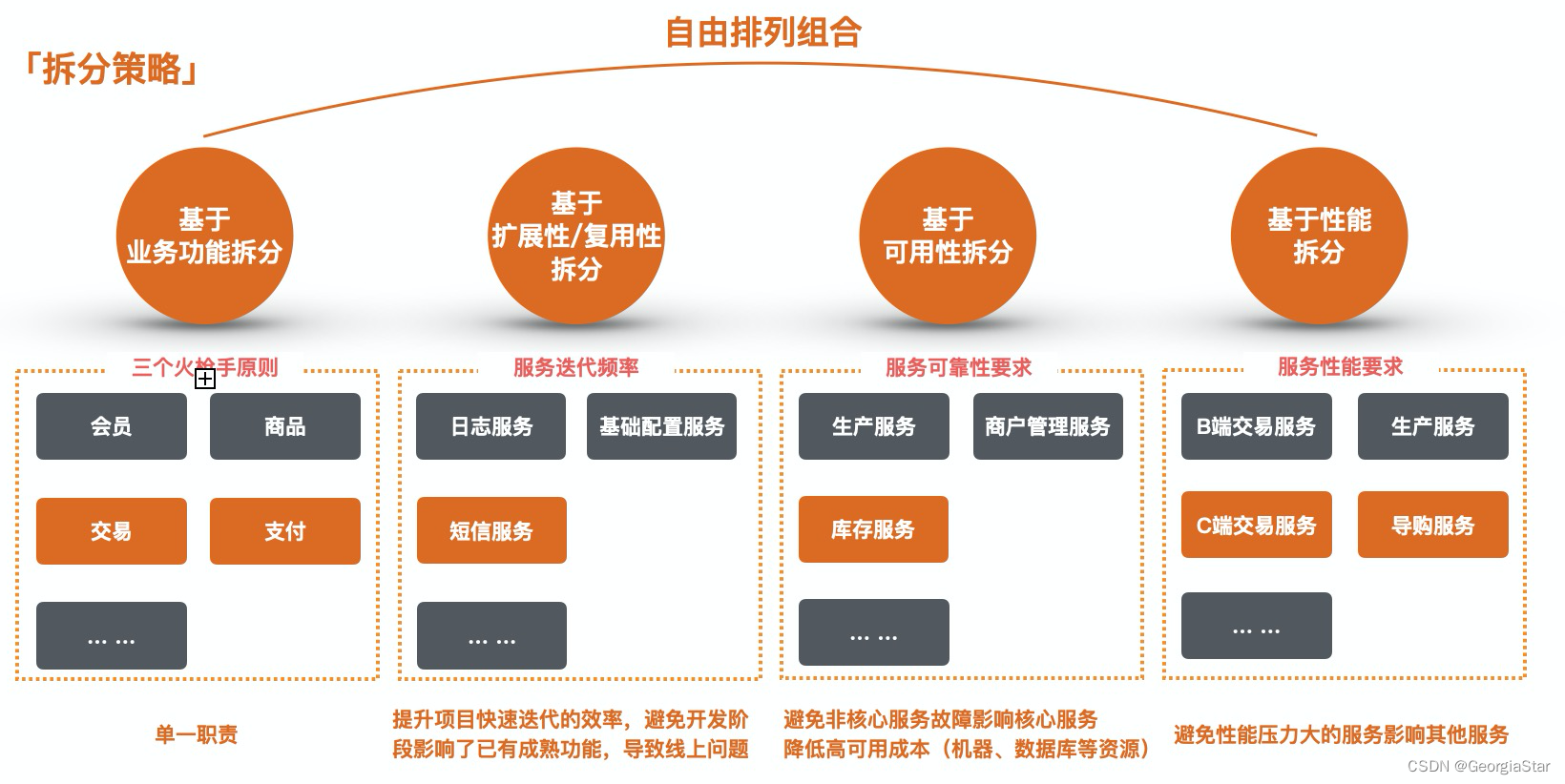
Microservice
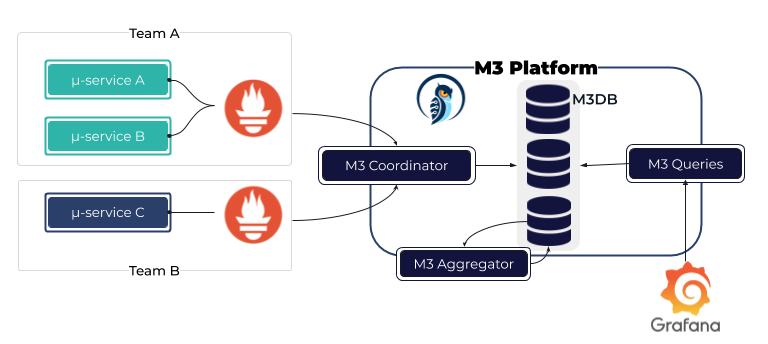
Observable time series data downsampling practice in Prometheus
![[IELTS reading] Wang Xiwei reads P4 (matching2 paragraph information matching question [difficult])](/img/83/63296108b47eda37c19b9ff9deb5ec.png)
[IELTS reading] Wang Xiwei reads P4 (matching2 paragraph information matching question [difficult])
随机推荐
What is the difference between port mapping and port forwarding
【雅思阅读】王希伟阅读P3(Heading)
微软禁用IE浏览器后,打开IE浏览器闪退解决办法
Robot reinforcement learning synergies between pushing and grassing with self supervised DRL (2018)
公司要上监控,Zabbix 和 Prometheus 怎么选?这么选准没错!
S32 design studio for arm 2.2 quick start
"Xiaodeng" domain password policy enhancer in operation and maintenance
微服务(Microservice)那点事儿
How to use fast parsing to make IOT cloud platform
How to save your code works quickly to better protect your labor achievements
Using fast parsing intranet penetration to realize zero cost self built website
[path planning] RRT adds dynamic model for trajectory planning
Hash table, hash function, bloom filter, consistency hash
端口映射和端口转发区别是什么
Pytoch --- use pytoch to realize linknet for semantic segmentation
Go step on the pit - no required module provides package: go mod file not found in current directory or any parent
Is the account opening link of Huatai Securities with low commission safe?
跨域请求
图解网络:什么是网关负载均衡协议GLBP?
A mining of edu certificate station